3T7U
 
 | | A NeW Crystal structure of APC-ARM | | Descriptor: | Adenomatous polyposis coli protein, PHOSPHATE ION | | Authors: | Zhang, Z, Wu, G. | | Deposit date: | 2011-07-31 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the armadillo repeat domain of adenomatous polyposis coli which reveals its inherent flexibility
Biochem.Biophys.Res.Commun., 412, 2011
|
|
5HL3
 
 | | Crystal structure of Listeria monocytogenes InlP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lmo2470 protein | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Kiryukhina, O, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
8HGR
 
 | | The apo-flavodoxin monomer from Synechococcus elongatus PCC 7942 | | Descriptor: | CHLORIDE ION, Flavodoxin, MAGNESIUM ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
8HGQ
 
 | | The apo-flavodoxin dimer from Synechococcus elongatus PCC 7942 | | Descriptor: | Flavodoxin, PHOSPHATE ION | | Authors: | Liu, S.W, Chen, Y.Y, Gong, Y, Cao, P. | | Deposit date: | 2022-11-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A dimer-monomer transition captured by the crystal structures of cyanobacterial apo flavodoxin.
Biochem.Biophys.Res.Commun., 639, 2022
|
|
3VBG
 
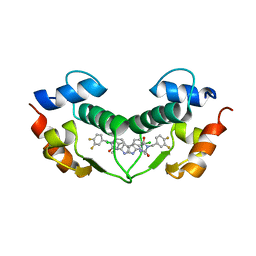 | | Structure of hDM2 with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2012-01-02 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3U15
 
 | | Structure of hDMX with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, Protein Mdm4, SULFATE ION | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7YAV
 
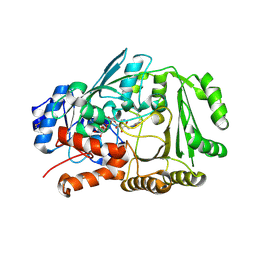 | | Crystal structure of Diels-Alderase MaDA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MaDA1 | | Authors: | Lei, X.G, Chen, R.C, Du, X.X, Yang, J, Fan, J.P, Guo, N.X, Ding, Q. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba.
Nat Commun, 15, 2024
|
|
7YOC
 
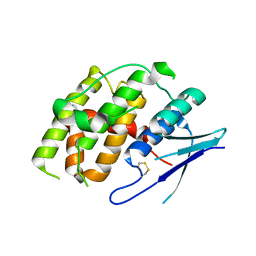 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YP0
 
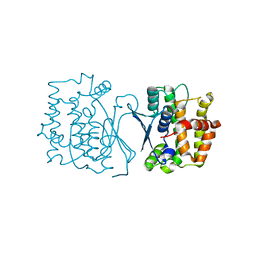 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7WEX
 
 | |
4J3Y
 
 | | Crystal structure of XIAP-BIR2 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J48
 
 | | Crystal structure of XIAP-BIR2 domain with AMRV bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, GLYCEROL, PEPTIDE (ALA-MET-ARG-VAL), ... | | Authors: | Gosu, R. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J46
 
 | | Crystal structure of XIAP-BIR2 domain with AVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J44
 
 | | Crystal structure of XIAP-BIR2 domain with AIAV bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-ILE-ALA-VAL), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J45
 
 | | Crystal structure of XIAP-BIR2 domain with ATAA bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-THR-ALA-ALA), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KJU
 
 | | Crystal structure of XIAP-Bir2 with a bound benzodiazepinone inhibitor. | | Descriptor: | E3 ubiquitin-protein ligase XIAP, N-{(3S)-5-(4-aminobenzoyl)-1-[(2-methoxynaphthalen-1-yl)methyl]-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepin-3-yl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
4J47
 
 | | Crystal structure of XIAP-BIR2 domain with SVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (SER-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MEJ
 
 | | Crystal structure of Lactobacillus helveticus purine deoxyribosyl transferase (PDT) with the tricyclic purine 8,9-dihydro-9-oxoimidazo[2,1-b]purine (N2,3-ethenoguanine) | | Descriptor: | 3H-imidazo[2,1-b]purin-4(5H)-one, Nucleoside deoxyribosyltransferase, SULFATE ION | | Authors: | Paul, D, Seckute, J, Ealick, S.E. | | Deposit date: | 2013-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosylation of a tricyclic purine analog at alternative
sites by nucleoside 2 -deoxyribosyltransferases
Plos One, 2014
|
|
4KJV
 
 | | Crystal structure of XIAP-Bir2 with a bound spirocyclic benzoxazepinone inhibitor. | | Descriptor: | 6-methoxy-5-({(3S)-3-[(N-methyl-L-alanyl)amino]-4-oxo-2',3,3',4,5',6'-hexahydro-5H-spiro[1,5-benzoxazepine-2,4'-pyran]-5-yl}methyl)naphthalene-2-carboxylic acid, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
5HZ6
 
 | |
5HZ8
 
 | | FABP4_3 in complex with 6,8-Dichloro-4-phenyl-2-piperidin-1-yl-quinoline-3-carboxylic acid | | Descriptor: | 6,8-dichloro-4-phenyl-2-(piperidin-1-yl)quinoline-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Rudolph, M.G. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Design and synthesis of selective, dual fatty acid binding protein 4 and 5 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5HZ9
 
 | |
5HZ5
 
 | |
7E9Q
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9O
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
