5WDT
 
 | | 70S ribosome-EF-Tu H84A complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WE6
 
 | | 70S ribosome-EF-Tu H84A complex with GTP and cognate tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
3DG2
 
 | |
3DG5
 
 | |
1ZN0
 
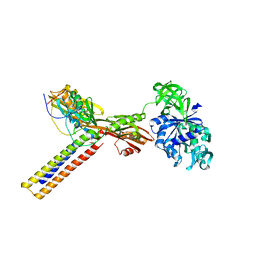 | | Coordinates of RRF and EF-G fitted into Cryo-EM map of the 50S subunit bound with both EF-G (GDPNP) and RRF | | Descriptor: | 16S RIBOSOMAL RNA, ELONGATION FACTOR G, Ribosome recycling factor | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
8G38
 
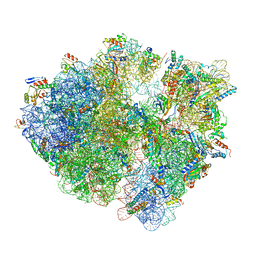 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
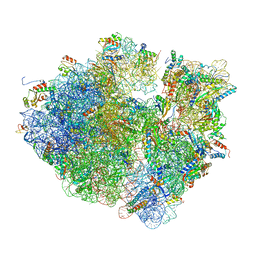 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
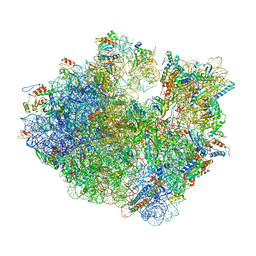 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
6XE0
 
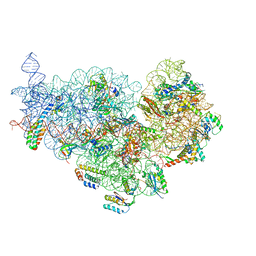 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
6PV6
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2019-07-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
1TRJ
 
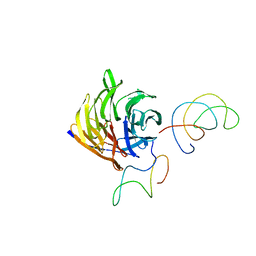 | | Homology Model of Yeast RACK1 Protein fitted into 11.7A cryo-EM map of Yeast 80S Ribosome | | Descriptor: | Guanine nucleotide-binding protein beta subunit-like protein, Helix 39 of 18S rRNA, Helix 40 of 18S rRNA | | Authors: | Sengupta, J, Nilsson, J, Gursky, R, Spahn, C.M, Nissen, P, Frank, J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM
Nat.Struct.Mol.Biol., 11, 2004
|
|
1JQT
 
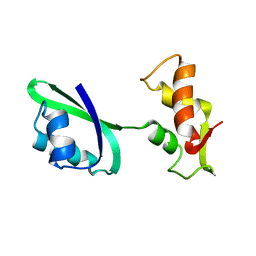 | | Fitting of L11 protein in the low resolution cryo-EM map of E.coli 70S ribosome | | Descriptor: | 50S Ribosomal protein L11 | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQM
 
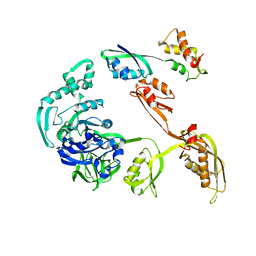 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQS
 
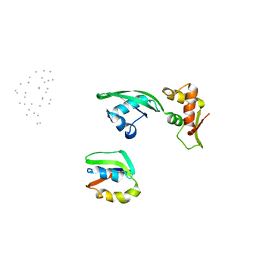 | | Fitting of L11 protein and elongation factor G (domain G' and V) in the cryo-em map of E. coli 70S ribosome bound with EF-G and GMPPCP, a nonhydrolysable GTP analog | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1LS2
 
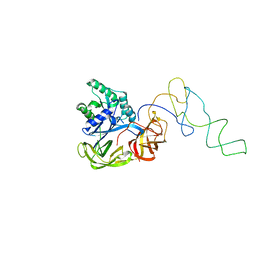 | | Fitting of EF-Tu and tRNA in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | Elongation Factor Tu, Phenylalanine transfer RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-16 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
1LU3
 
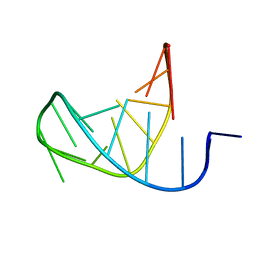 | | Separate Fitting of the Anticodon Loop Region of tRNA (nucleotide 26-42) in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | Descriptor: | PHENYLALANINE TRANSFER RNA | | Authors: | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16.799999 Å) | | Cite: | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
1MI6
 
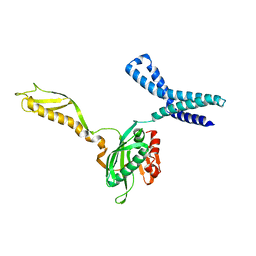 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | Descriptor: | peptide chain release factor RF-2 | | Authors: | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
1MVR
 
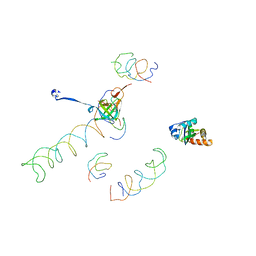 | | Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome | | Descriptor: | 30S RIBOSOMAL PROTEIN S12, 50S ribosomal protein L11, Helix 34 of 16S rRNA, ... | | Authors: | Rawat, U.B, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
5KBS
 
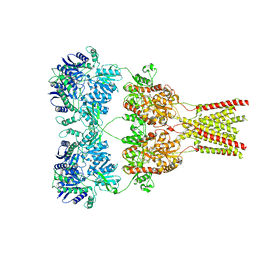 | | Cryo-EM structure of GluA2-0xSTZ at 8.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBV
 
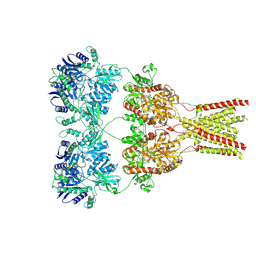 | | Cryo-EM structure of GluA2 bound to antagonist ZK200775 at 6.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.G, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBU
 
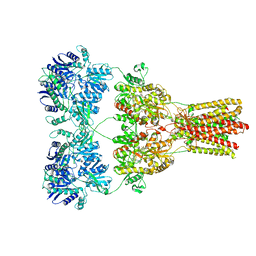 | | Cryo-EM structure of GluA2-2xSTZ complex at 7.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBT
 
 | | Cryo-EM structure of GluA2-1xSTZ complex at 6.4 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
