2BTL
 
 | |
2BXX
 
 | |
7YRV
 
 | |
7YRW
 
 | |
7YRX
 
 | |
6U6T
 
 | | Neuronal growth regulator 1 (NEGR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuronal growth regulator 1, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
7X4Y
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GABA complex | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X52
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP complex | | Descriptor: | ACETATE ION, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X51
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GUA complex | | Descriptor: | GLUTARIC ACID, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, L, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X4L
 
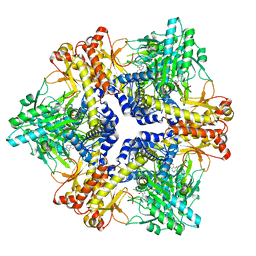 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | Deposit date: | 2022-03-02 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
6M0J
 
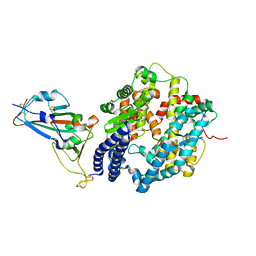 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
4RP9
 
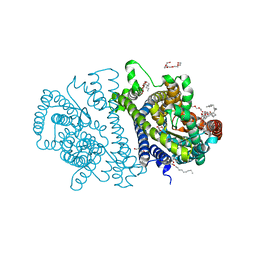 | | Bacterial vitamin C transporter UlaA/SgaT in C2 form | | Descriptor: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, PENTAETHYLENE GLYCOL, ... | | Authors: | Wang, J.W. | | Deposit date: | 2014-10-29 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RP8
 
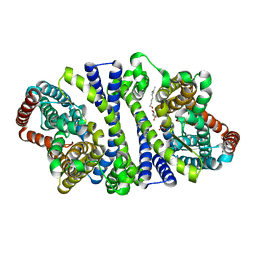 | | Bacterial vitamin C transporter UlaA/SgaT in P21 form | | Descriptor: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, nonyl beta-D-glucopyranoside | | Authors: | Wang, J.W. | | Deposit date: | 2014-10-29 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5J9U
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9T
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9Q
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9W
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | ACETYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
6U7N
 
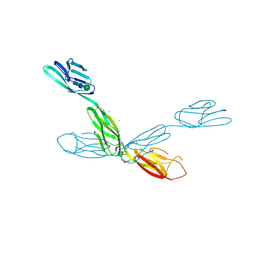 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
6LJH
 
 | | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua in complex with NAD+ | | Descriptor: | Alcohol dehydrogenase 1, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Feng, X, Fan, S, Li, M, Zou, S, Lv, G, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2019-12-16 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua in complex with NAD+
To Be Published
|
|
6BOZ
 
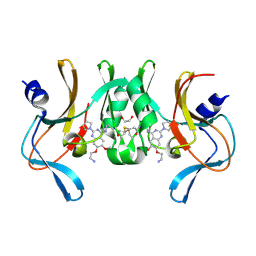 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
7KXT
 
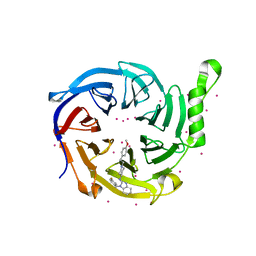 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
5HYW
 
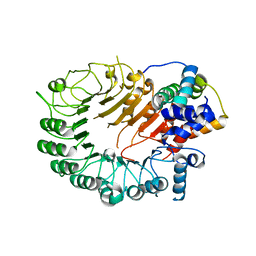 | | The crystal structure of the D3-ASK1 complex | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
5HZG
 
 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
5V2N
 
 | | Crystal Structure of APO Human SETD8 | | Descriptor: | 1,2-ETHANEDIOL, N-lysine methyltransferase KMT5A | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-05 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
5YIM
 
 | | Structure of a Legionella effector | | Descriptor: | SdeA | | Authors: | Feng, Y, Dong, Y, Wang, W. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
