5UMV
 
 | |
5VF0
 
 | |
5TAB
 
 | |
8S9K
 
 | | Structure of dimeric FAM111A SPD S541A Mutant | | Descriptor: | GLYCEROL, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
8S9L
 
 | | Structure of monomeric FAM111A SPD V347D Mutant | | Descriptor: | SULFATE ION, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
3FSS
 
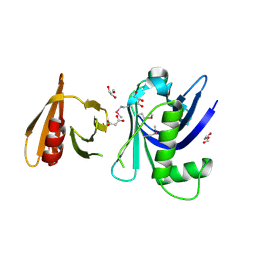 | | Structure of the tandem PH domains of Rtt106 | | Descriptor: | GLYCEROL, Histone chaperone RTT106, MALONIC ACID | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2009-01-11 | | Release date: | 2009-12-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
6OW7
 
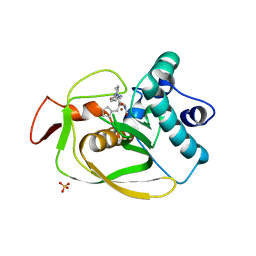 | | X-ray Structure of Polypeptide Deformylase with a Piperazic Acid | | Descriptor: | (3S)-2-{(2R)-2-(cyclopentylmethyl)-3-[formyl(hydroxy)amino]propanoyl}-N-(pyridin-2-yl)hexahydropyridazine-3-carboxamide, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8U3S
 
 | |
8UQE
 
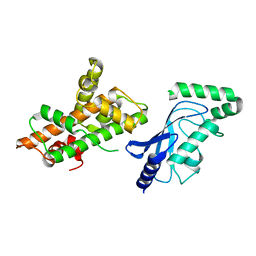 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.562 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQA
 
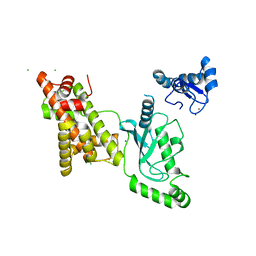 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 12-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, SODIUM ION, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQB
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ9
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 4-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ8
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 2-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQD
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.893 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQC
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 2) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
6OW2
 
 | | X-ray Structure of Polypeptide Deformylase | | Descriptor: | (2R)-2-(cyclopentylmethyl)-N'-{5-fluoro-6-[(9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]-2-methylpyrimidin-4-yl}-3-[hydroxy(hydroxymethyl)amino]propanehydrazide, NICKEL (II) ION, Peptide deformylase | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4OEM
 
 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
4OEL
 
 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Dipeptidyl peptidase 1 Heavy chain, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
7WAT
 
 | |
3TVV
 
 | |
3TW1
 
 | | Structure of Rtt106-AHN | | Descriptor: | GLYCEROL, Histone chaperone RTT106, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
