8VUF
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5i3 | | Descriptor: | (2E)-3-[4-({2-[(1-{[4-(methanesulfonyl)phenyl]methyl}piperidin-4-yl)amino]pyrido[2,3-d]pyrimidin-4-yl}oxy)-3,5-dimethylphenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
8VUB
 
 | | Crystal structure of wild-type HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
8VUM
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
6KOF
 
 | | Crystal structure of indoleamine 2,3-dioxygenagse 1 (IDO1) in complex with compound 47 | | Descriptor: | 1-(4-cyanophenyl)-3-[[3-(2-cyclopropylethynyl)imidazo[2,1-b][1,3]thiazol-5-yl]methyl]thiourea, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Peng, Y.H, Wu, S.Y. | | Deposit date: | 2019-08-09 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Unique Sulfur-Aromatic Interactions Contribute to the Binding of Potent Imidazothiazole Indoleamine 2,3-Dioxygenase Inhibitors.
J.Med.Chem., 63, 2020
|
|
8GMN
 
 | | Crystal structure of human C1s in complex with inhibitor | | Descriptor: | Complement C1s subcomponent, SULFATE ION, [4-(1-aminophthalazin-6-yl)piperazin-1-yl](2-methylphenyl)methanone | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2023-03-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway.
J.Med.Chem., 66, 2023
|
|
6E3E
 
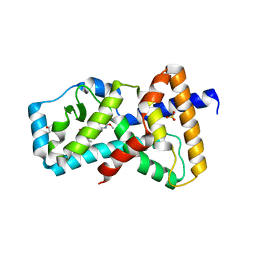 | | Structure of RORgt in complex with a novel inverse agonist. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(5R)-5-[(7-fluoro-1,1-dimethyl-2,3-dihydro-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5H)-yl]-5-oxopentanoic acid, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
6E3G
 
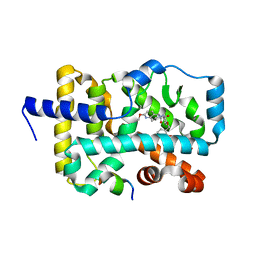 | | Structure of RORgt in complex with a novel agonist. | | Descriptor: | (5R)-6-acetyl-2-methoxy-N-{4-[(2-methoxyphenyl)methoxy]phenyl}-5,6,7,8-tetrahydro-1,6-naphthyridine-5-carboxamide, 1,2-ETHANEDIOL, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
7DUT
 
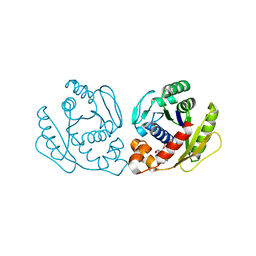 | | Structure of Sulfolobus solfataricus SegA protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DUV
 
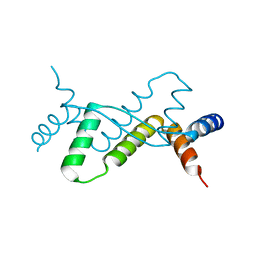 | | Structure of Sulfolobus solfataricus SegB protein | | Descriptor: | SULFATE ION, SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV2
 
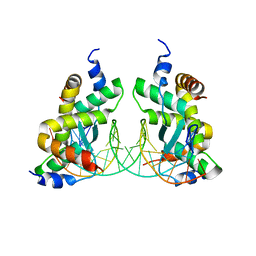 | | Structure of Sulfolobus solfataricus SegB-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*AP*GP*AP*AP*GP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*CP*TP*TP*CP*TP*AP*CP*GP*TP*A)-3'), SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV3
 
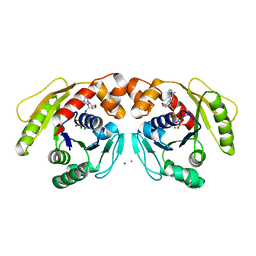 | | Structure of Sulfolobus solfataricus SegA-AMPPNP protein | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Wu, C.T, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DWR
 
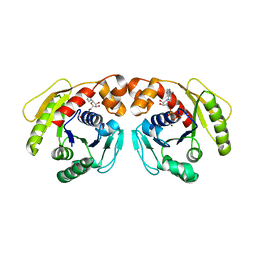 | | Structure of Sulfolobus solfataricus SegA-ADP complex bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
1KC7
 
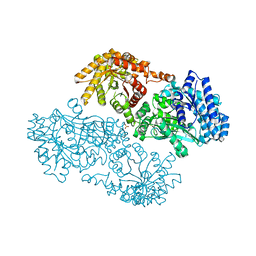 | | Pyruvate Phosphate Dikinase with Bound Mg-phosphonopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHONOPYRUVATE, SULFATE ION, ... | | Authors: | Chen, C.C, Howard, A, Herzberg, O. | | Deposit date: | 2001-11-07 | | Release date: | 2002-01-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvate site of pyruvate phosphate dikinase: crystal structure of the enzyme-phosphonopyruvate complex, and mutant analysis
Biochemistry, 41, 2002
|
|
1DIK
 
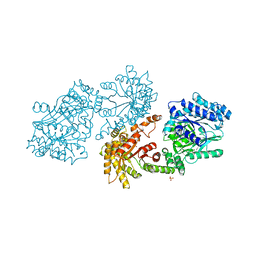 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3RH8
 
 | |
1C8A
 
 | |
5VG1
 
 | |
5VG0
 
 | |
1GPR
 
 | |
1C89
 
 | |
5UJK
 
 | |
5ULV
 
 | | Malate dehydrogenase from Methylobacterium extorquens | | Descriptor: | CALCIUM ION, Malate dehydrogenase | | Authors: | Gonzalez, J.M. | | Deposit date: | 2017-01-25 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational changes on substrate binding revealed by structures of Methylobacterium extorquens malate dehydrogenase.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4ROP
 
 | |
3NLA
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3RDN
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
