6KVC
 
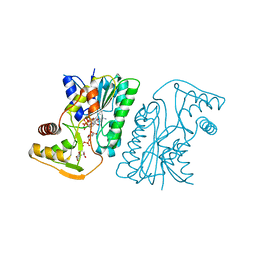 | | MoeE5 in complex with UDP-glucose and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
7XTV
 
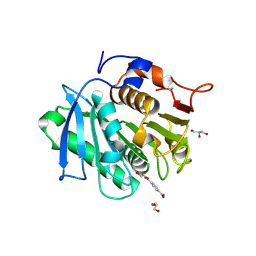 | | The structure of MHET-bound TfCut S130A | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTU
 
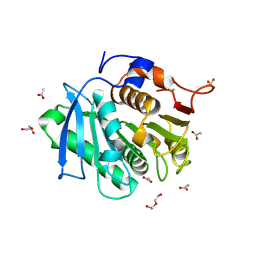 | | The structure of TfCut S130A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTS
 
 | | The apo structure of the engineered TfCut S130A | | Descriptor: | SODIUM ION, alpha/beta hydrolase | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTW
 
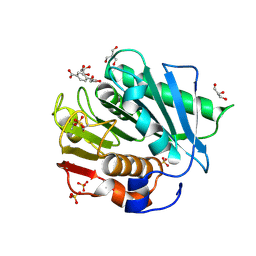 | | The structure of IsPETase in complex with MHET | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTR
 
 | | The apo structure of the engineered TfCut | | Descriptor: | GLYCEROL, LITHIUM ION, SULFATE ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTT
 
 | | The structure of engineered TfCut S130A in complex with MHET | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, SODIUM ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7W1W
 
 | | NADPH-bound AKR4C17 mutant F291D | | Descriptor: | AKR4-2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-11-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7W1X
 
 | | Crystal structure of AKR4C16 bound with NADPH | | Descriptor: | AKR4-1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-11-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7FIR
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with 1,4-mannobiose | | Descriptor: | Beta-1,2-mannobiose phosphorylase, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIP
 
 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIS
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with mannose 1-phosphate (M1P) | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, Beta-1,2-mannobiose phosphorylase, GLYCEROL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7F7M
 
 | | AKR4C17 in complex with NADP+ and glyphosate | | Descriptor: | AKR4-2, GLYPHOSATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7L
 
 | | Crystal structure of AKR4C17 bound with NADPH | | Descriptor: | AKR4-2, COBALT (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7K
 
 | | Crystal structure of AKR4C17 bound with NADP+ | | Descriptor: | AKR4-2, COBALT (II) ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7J
 
 | | The crystal structure of AKR4C17 | | Descriptor: | AKR4-2, COBALT (II) ION, SULFATE ION | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7EWA
 
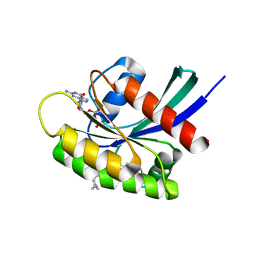 | | GDP-bound KRAS G12D in complex with TH-Z827 | | Descriptor: | 4-[(1~{R},5~{S})-3,8-diazabicyclo[3.2.1]octan-8-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
7EW9
 
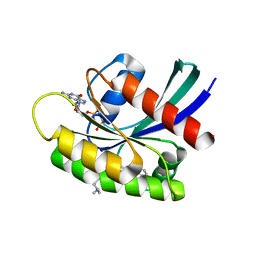 | | GDP-bound KRAS G12D in complex with TH-Z816 | | Descriptor: | 7-(8-methylnaphthalen-1-yl)-4-[(2~{R})-2-methylpiperazin-1-yl]-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
7EWB
 
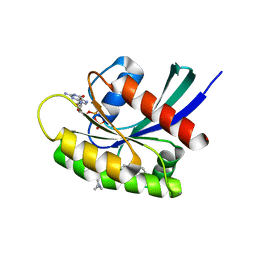 | | GDP-bound KRAS G12D in complex with TH-Z835 | | Descriptor: | 4-[(1~{S},5~{R})-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
