6G1J
 
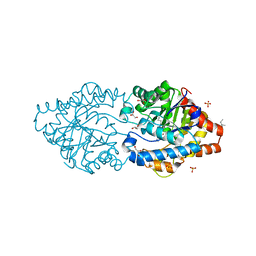 | | Phosphotriesterase PTE_C23M_1 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphotriesterase
PTE_C23M_1
To Be Published
|
|
6FWE
 
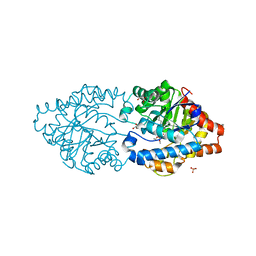 | | Phosphotriesterase PTE_C23_6 | | Descriptor: | 1-[methoxy(methyl)phosphoryl]oxyethane, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Phosphotriesterase
PTE_C23_6
To Be Published
|
|
6GC2
 
 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
6FRZ
 
 | | Phosphotriesterase PTE_A53_7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methylcyclohexane-1,1,3,3-tetrol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphotriesterase
PTE_A53_7
To Be Published
|
|
6G3M
 
 | | Phosphotriesterase PTE_C23M_4 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Phosphotriesterase
PTE_C23M_4
To Be Published
|
|
2GZU
 
 | |
2MF7
 
 | | Solution structure of the ims domain of the mitochondrial import protein TIM21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bajaj, R, Jaremko, L, Jaremko, M, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of the Dynamic Structure of the TIM23 Complex in the Mitochondrial Intermembrane Space.
Structure, 22, 2014
|
|
8B1P
 
 | | Crystal structure of SUDV VP40 CCS mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
8B3X
 
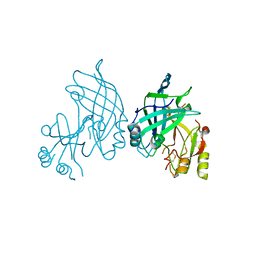 | | High resolution crystal structure of dimeric SUDV VP40 | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Norris, M, Saphire, E.O, Becker, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
8B1O
 
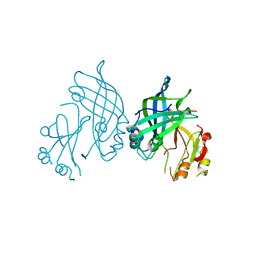 | | Crystal structure of SUDV VP40 C314S mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
2KOX
 
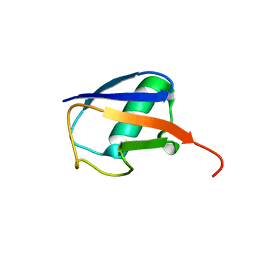 | | NMR residual dipolar couplings identify long range correlated motions in the backbone of the protein ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Fenwick, R.B, Richter, B, Lee, D, Walter, K.F.A, Milovanovic, D, Becker, S, Lakomek, N.A, Griesinger, C, Salvatella, X. | | Deposit date: | 2009-10-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Weak Long-Range Correlated Motions in a Surface Patch of Ubiquitin Involved in Molecular Recognition
J.Am.Chem.Soc., 2011
|
|
1OJ5
 
 | | Crystal structure of the Nco-A1 PAS-B domain bound to the STAT6 transactivation domain LXXLL motif | | Descriptor: | IODIDE ION, SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 6, STEROID RECEPTOR COACTIVATOR 1A | | Authors: | Razeto, A, Ramakrishnan, V, Giller, K, Lakomek, N, Carlomagno, T, Griesinger, C, Lodrini, M, Litterst, C.M, Pftizner, E, Becker, S. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Ncoa-1/Src-1 Pas-B Domain Bound to the Lxxll Motif of the Stat6 Transactivation Domain
J.Mol.Biol., 336, 2004
|
|
5HQ3
 
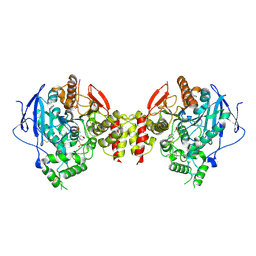 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
4C4Z
 
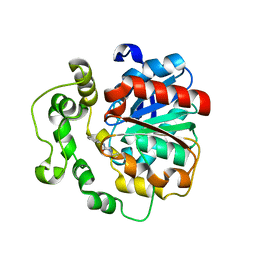 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A8 | | Descriptor: | 1-ethyl-3-naphthalen-1-ylurea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4Y
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with A4 | | Descriptor: | 1-(3-chlorophenyl)-3-(2-methoxyethyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C4X
 
 | | Crystal structure of human bifunctional epoxide hydroxylase 2 complexed with C9 | | Descriptor: | 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2 | | Authors: | Pilger, J, Mazur, A, Monecke, P, Schreuder, H, Elshorst, B, Langer, T, Schiffer, A, Krimm, I, Wegstroth, M, Lee, D, Hessler, G, Wendt, K.-U, Becker, S, Griesinger, C. | | Deposit date: | 2013-09-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Combination of Spin Diffusion Methods for the Determination of Protein-Ligand Complex Structural Ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2MME
 
 | | Hybrid structure of the Shigella flexneri MxiH Type three secretion system needle | | Descriptor: | MxiH | | Authors: | Demers, J.P, Habenstein, B, Loquet, A, Vasa, S.K, Becker, S, Baker, D, Lange, A, Sgourakis, N.G. | | Deposit date: | 2014-03-14 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.7 Å), SOLID-STATE NMR | | Cite: | High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy.
Nat Commun, 5, 2014
|
|
3EE6
 
 | | Crystal Structure Analysis of Tripeptidyl peptidase -I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pal, A, Kraetzner, R, Grapp, M, Gruene, T, Schreiber, K, Granborg, M, Urlaub, H, Asif, A.R, Becker, S, Gartner, J, Sheldrick, G.M, Steinfeld, R. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
3HQJ
 
 | | Structure-function analysis of Mycobacterium tuberculosis acyl carrier protein synthase (AcpS). | | Descriptor: | COENZYME A, Holo-[acyl-carrier-protein] synthase, MAGNESIUM ION | | Authors: | Dym, O, Albeck, S, Peleg, Y, Schwarz, A, Shakked, Z, Burstein, Y, Zimhony, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-06-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function analysis of the acyl carrier protein synthase (AcpS) from Mycobacterium tuberculosis.
J.Mol.Biol., 393, 2009
|
|
8OWD
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L1 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWJ
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVM
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWE
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
