6TUV
 
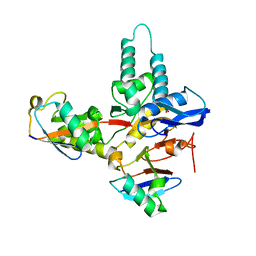 | |
6T68
 
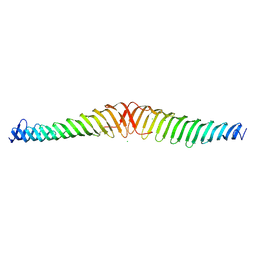 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6ENR
 
 | |
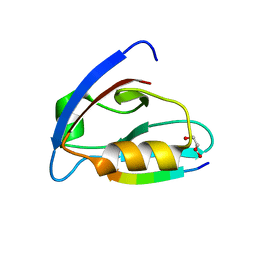
6EZI
 
 | |
6FKG
 
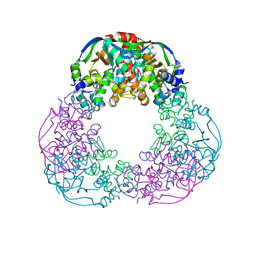 | | Crystal structure of the M.tuberculosis MbcT-MbcA toxin-antitoxin complex. | | Descriptor: | GLYCEROL, Rv1989c (MbcT), Rv1990c (MbcA) | | Authors: | Freire, D.M, Cianci, M, Pogenberg, V, Schneider, T.R, Wilmanns, M, Parret, A.H.A. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An NAD+Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death.
Mol.Cell, 73, 2019
|
|
6FKQ
 
 | | THE CRYSTAL STRUCTURE OF A FRAGMENT OF NETRIN-1 IN COMPLEX WITH A FRAGMENT OF DRAXIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bhowmick, T, Meijers, R. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
6FHB
 
 | |
6FHA
 
 | |
4LZ2
 
 | | Crystal structure of the bromodomain of human BAZ2A | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION | | Authors: | Tallant, C, Nunez-Alonso, G, Picaud, S, Filippakopoulos, P, Krojer, T, Bradley, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
4MB4
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB3
 
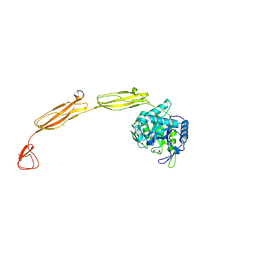 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NK7
 
 | |
4MB5
 
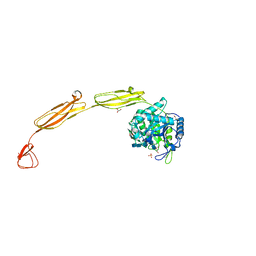 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ADF
 
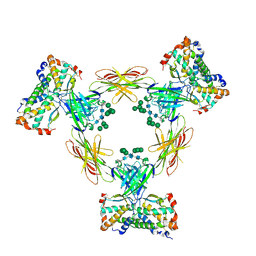 | | CRYSTAL STRUCTURE OF THE HUMAN COLONY-STIMULATING FACTOR 1 (hCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 | | Descriptor: | MACROPHAGE COLONY-STIMULATING FACTOR 1, SECRETED PROTEIN BARF1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Bracke, N, Savvides, S.N. | | Deposit date: | 2011-12-23 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Allosteric Competitive Inactivation of Hematopoietic Csf-1 Signaling by the Viral Decoy Receptor Barf1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4ADQ
 
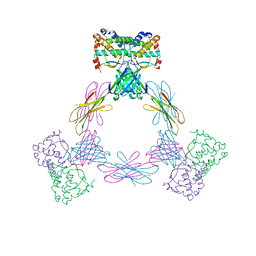 | | CRYSTAL STRUCTURE OF THE MOUSE COLONY-STIMULATING FACTOR 1 (MCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 | | Descriptor: | MACROPHAGE COLONY-STIMULATING FACTOR 1, SECRETED PROTEIN BARF1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Bracke, N, Savvides, S.N. | | Deposit date: | 2012-01-02 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Allosteric Competitive Inactivation of Hematopoietic Csf-1 Signaling by the Viral Decoy Receptor Barf1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4OW2
 
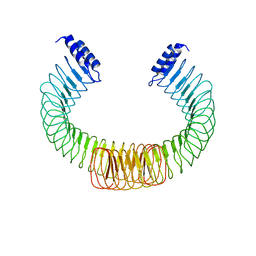 | | YopM from Yersinia enterocolitica WA-314 | | Descriptor: | Yop effector YopM | | Authors: | Rumm, A, Perbandt, M, Aepfelbacher, M. | | Deposit date: | 2014-01-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog., 12, 2016
|
|
4AQP
 
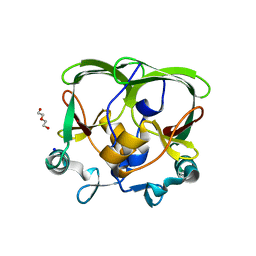 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
4APT
 
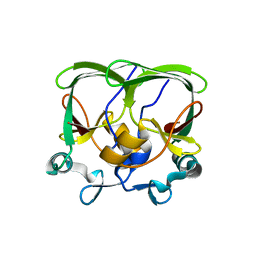 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
4NKB
 
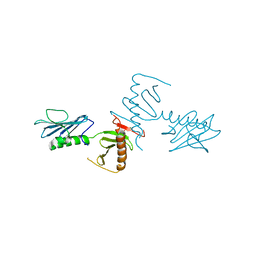 | | Crystal Structure of the cryptic polo box (CPB)of ZYG-1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Shimanovskaya, E, Dong, G. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C. elegans ZYG-1 Cryptic Polo Box Suggests a Conserved Mechanism for Centriolar Docking of Plk4 Kinases.
Structure, 22, 2014
|
|
3UTO
 
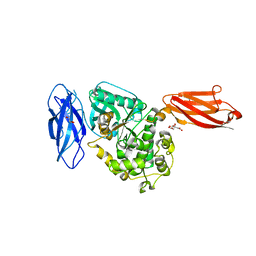 | | Twitchin kinase region from C.elegans (Fn31-NL-kin-CRD-Ig26) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Castelmur, E, Barbieri, S, Mayans, O. | | Deposit date: | 2011-11-26 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of an N-terminal inhibitory extension as the primary mechanosensory regulator of twitchin kinase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1OFF
 
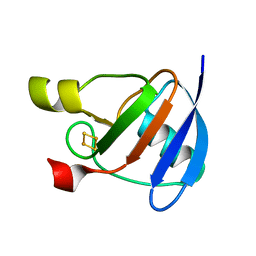 | |
1OFD
 
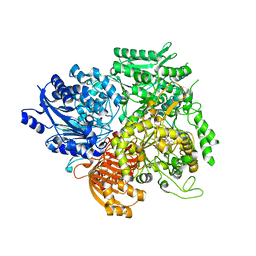 | |
1OFE
 
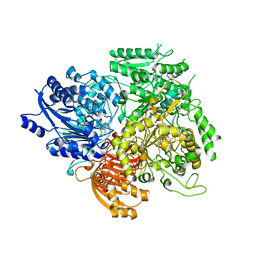 | |
4V48
 
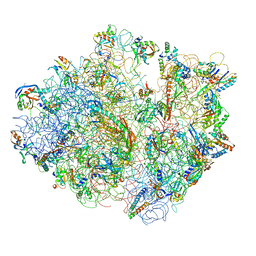 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
2HAK
 
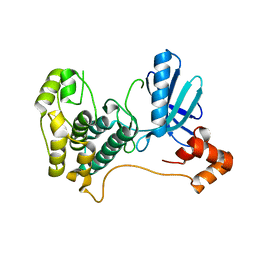 | | Catalytic and ubiqutin-associated domains of MARK1/PAR-1 | | Descriptor: | Serine/threonine-protein kinase MARK1 | | Authors: | Marx, A, Nugoor, C, Mueller, J, Panneerselvam, S, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2
J.Biol.Chem., 281, 2006
|
|
