5VPB
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-I crystal | | Descriptor: | CHLORIDE ION, Protein fosB, Transcription factor jun-D | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
1JPZ
 
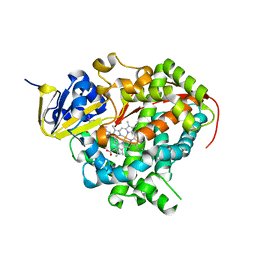 | | Crystal structure of a complex of the heme domain of P450BM-3 with N-Palmitoylglycine | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Haines, D.C, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pivotal role of water in the mechanism of P450BM-3.
Biochemistry, 40, 2001
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
1N11
 
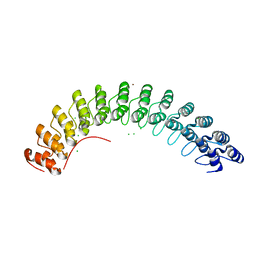 | | D34 REGION OF HUMAN ANKYRIN-R AND LINKER | | Descriptor: | Ankyrin, BROMIDE ION, CHLORIDE ION | | Authors: | Michaely, P, Tomchick, D.R, Machius, M, Anderson, R.G.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a 12 ANK repeat stack from human ankyrinR
Embo J., 21, 2002
|
|
1NHH
 
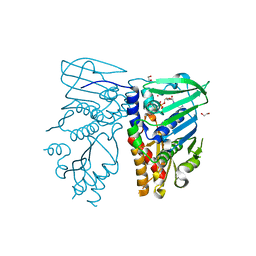 | | Crystal structure of N-terminal 40KD MutL protein (LN40) complex with ADPnP and one Rubidium | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
1NHJ
 
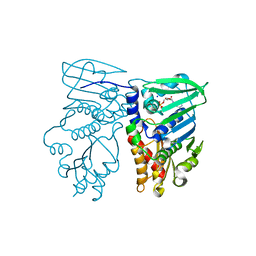 | | Crystal structure of N-terminal 40KD MutL/A100P mutant protein complex with ADPnP and one sodium | | Descriptor: | DNA mismatch repair protein mutL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
1NHI
 
 | | Crystal structure of N-terminal 40KD MutL (LN40) complex with ADPnP and one potassium | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
2Q3X
 
 | | The RIM1alpha C2B domain | | Descriptor: | CHLORIDE ION, Regulating synaptic membrane exocytosis protein 1, SODIUM ION, ... | | Authors: | Guan, R, Dai, H, Tomchick, D.R, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2007-05-30 | | Release date: | 2007-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the RIM1alpha C(2)B Domain at 1.7 A Resolution.
Biochemistry, 46, 2007
|
|
1OLS
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1OLU
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-15 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1O75
 
 | | Tp47, the 47-Kilodalton Lipoprotein of Treponema pallidum | | Descriptor: | 2,3-di-O-sulfo-alpha-D-glucopyranose-(1-6)-2,3-di-O-sulfo-alpha-D-glucopyranose, 47 KDA MEMBRANE ANTIGEN, XENON | | Authors: | Deka, R.K, Machius, M, Norgard, M.V, Tomchick, D.R. | | Deposit date: | 2002-10-23 | | Release date: | 2002-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the 47-kDa lipoprotein of Treponema pallidum reveals a novel penicillin-binding protein.
J. Biol. Chem., 277, 2002
|
|
1OLX
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-18 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
3V1A
 
 | | Crystal structure of de novo designed MID1-apo1 | | Descriptor: | Computational design, MID1-apo1 | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1E
 
 | | Crystal structure of de novo designed MID1-zinc H12E mutant | | Descriptor: | Computational design, MID1-zinc H12E mutant, ZINC ION | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.073 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1C
 
 | | Crystal structure of de novo designed MID1-zinc | | Descriptor: | Computational design, MID1-zinc, L(+)-TARTARIC ACID, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.129 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1F
 
 | | Crystal structure of de novo designed MID1-zinc H35E mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Computational design, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1D
 
 | | Crystal structure of de novo designed MID1-cobalt | | Descriptor: | COBALT (II) ION, Computational design, MID1-cobalt, ... | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3V1B
 
 | | Crystal structure of de novo designed MID1-apo2 | | Descriptor: | Computational design, MID1-apo2, GLYCEROL | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
3ZY7
 
 | | Crystal structure of computationally redesigned gamma-adaptin appendage domain forming a symmetric homodimer | | Descriptor: | AP-1 COMPLEX SUBUNIT GAMMA-1, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Stranges, P.B, Machius, M, Miley, M.J, Tripathy, A, Kuhlman, B. | | Deposit date: | 2011-08-17 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Computational Design of a Symmetric Homodimer Using Beta-Strand Assembly.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1U3C
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U5Q
 
 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | CALCIUM ION, serine/threonine protein kinase TAO2 | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
STRUCTURE, 12, 2004
|
|
1U3D
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana with AMPPNP bound | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U5R
 
 | | Crystal Structure of the TAO2 Kinase Domain: Activation and Specifity of a Ste20p MAP3K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhou, T, Raman, M, Gao, Y, Earnest, S, Chen, Z, Machius, M, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TAO2 Kinase Domain; Activation and Specificity of a Ste20p MAP3K.
Structure, 12, 2004
|
|
2OIV
 
 | | Structural Analysis of Xanthomonas XopD Provides Insights Into Substrate Specificity of Ubiquitin-like Protein Proteases | | Descriptor: | PHOSPHATE ION, Xanthomonas outer protein D | | Authors: | Chosed, R, Tomchick, D.R, Brautigam, C.A, Machius, M, Orth, K. | | Deposit date: | 2007-01-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases.
J.Biol.Chem., 282, 2007
|
|
