8ISP
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISO
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
4EDA
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
5C6F
 
 | | Crystal structures of ferritin mutants reveal side-on binding to diiron and end-on cleavage of oxygen | | Descriptor: | Bacterial non-heme ferritin, FE (III) ION, IMIDAZOLE | | Authors: | Kim, S, Kim, K.H, Seok, J.H, Park, Y.H, Jung, S.W, Chung, Y.B, Lee, D.B, Lee, J.H, Han, K.R, Cho, A.E, Lee, C, Chung, M.S. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
4NU2
 
 | | Crystal structure of an ice-binding protein (FfIBP) from the Antarctic bacterium, Flavobacterium frigoris PS1 | | Descriptor: | Antifreeze protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4EDB
 
 | | Structures of monomeric hemagglutinin and its complex with an Fab fragment of a neutralizing antibody that binds to H1 subtype influenza viruses: molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Hemagglutinin | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-03-27 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into structural diversity of influenza virus haemagglutinin
J.Gen.Virol., 94, 2013
|
|
4NU3
 
 | | Crystal structure of mFfIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | SODIUM ION, SULFATE ION, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YX3
 
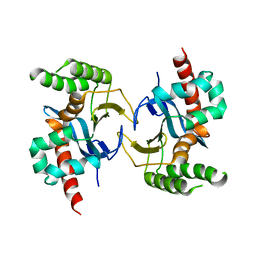 | |
4NUH
 
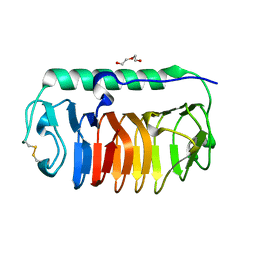 | | Crystal structure of mLeIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YX4
 
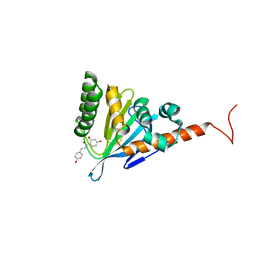 | | Isoliquiritigenin-complexed Chalcone isomerase (S189A) from the Antarctic vascular plant Deschampsia Antarctica (DaCHI1) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Chalcone-flavonone isomerase family protein | | Authors: | Lee, C.W, Park, S, Lee, J.H. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzymatic properties of chalcone isomerase from the Antarctic vascular plant Deschampsia antarctica Desv.
PLoS ONE, 13, 2018
|
|
5IT1
 
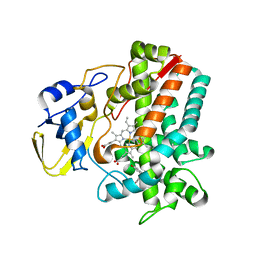 | | Streptomyces peucetius CYP105P2 complex with biphenyl compound | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2016-03-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytochrome P450 (CYP105P2) from Streptomyces peucetius and Its Conformational Changes in Response to Substrate Binding
Int J Mol Sci, 17, 2016
|
|
5XNT
 
 | | Structure of CYP106A2 from Bacillus sp. PAMC 23377 | | Descriptor: | Cytochrome P450 CYP106, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Kim, K.-H, Bikash, D, Park, S.-H, Park, H, Oh, T.-J, Lee, J.H. | | Deposit date: | 2017-05-24 | | Release date: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of a Cytochrome P450 (BaCYP106A2) fromBacillussp. PAMC 23377.
J. Microbiol. Biotechnol., 27, 2017
|
|
6A7J
 
 | | Testerone bound CYP154C4 from Streptomyces sp. ATCC 11861 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, TESTOSTERONE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
6A4D
 
 | |
6A4E
 
 | |
6A7I
 
 | | CYP154C4 from Streptomyces sp. W2061 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
6A4A
 
 | |
4I39
 
 | | Structures of ICT and PR1 intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I3A
 
 | | Structures of PR1 and PR2 intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4HY8
 
 | | Structures of PR1 and PR2 intermediates from time-resolved laue crystallography | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Wulff, M, Moffat, K. | | Deposit date: | 2012-11-13 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I38
 
 | | Structures of IT intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I3I
 
 | | Structures of IT intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected at 14ID APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I3J
 
 | | Structures of PR1 intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected AT 14ID APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
