5DSK
 
 | |
5DSQ
 
 | |
5DSM
 
 | |
5DSR
 
 | |
4XG8
 
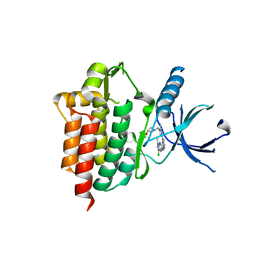 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(1-{2-[(3-chloro-1-methyl-1H-indazol-5-yl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
5GHV
 
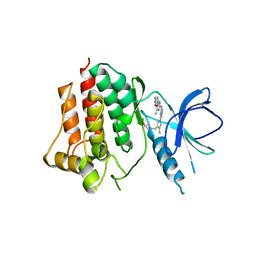 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-({1-[2-({3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]-4-methyl-1H-pyrrol-3-yl}methyl)azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
5DSP
 
 | |
7PQW
 
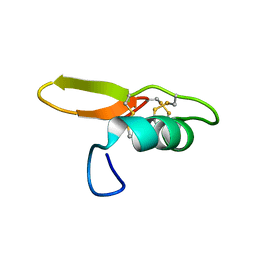 | | NMR solution structure of BCR4 | | Descriptor: | BCR4 | | Authors: | Loth, K, Paquet, F. | | Deposit date: | 2021-09-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Aphid BCR4 Structure and Activity Uncover a New Defensin Peptide Superfamily.
Int J Mol Sci, 23, 2022
|
|
6KUU
 
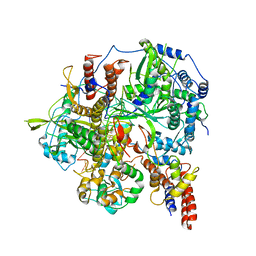 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
5F9W
 
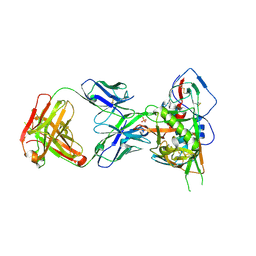 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of CH235-lineage antibody CH235, Light chain of CH235-lineage antibody CH235, ... | | Authors: | Chen, L, Zhou, T, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8911 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
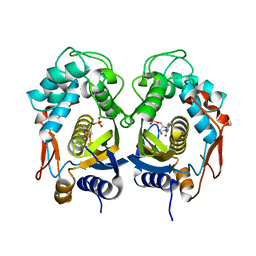
5WRN
 
 | |
5F96
 
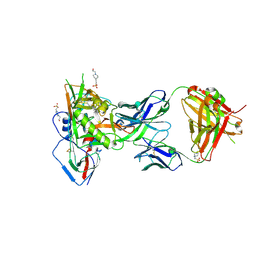 | |
8BVB
 
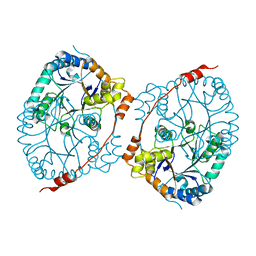 | |
8QFZ
 
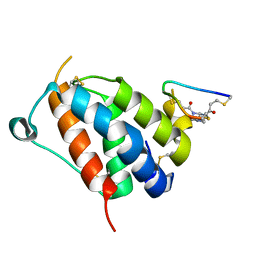 | | TSLP-Bicycle complex | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, CYS-HIS-TRP-LEU-GLU-ASN-CYS-TRP-ARG-GLY-PHE-CYS, Thymic stromal lymphopoietin | | Authors: | Petersen, J. | | Deposit date: | 2023-09-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Characterization of a Bicyclic Peptide (Bicycle) Binder to Thymic Stromal Lymphopoietin.
J.Med.Chem., 67, 2024
|
|
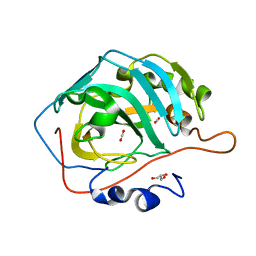
5DSO
 
 | |
5YUK
 
 | |
5YUI
 
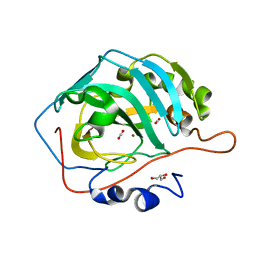 | | CO2 release in human carbonic anhydrase II crystals: reveal histidine 64 and solvent dynamics | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Park, S.Y, McKenna, R. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5F9O
 
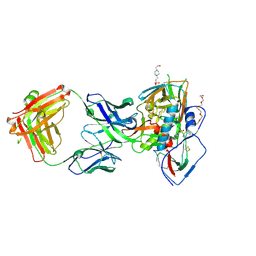 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5YUJ
 
 | |
5Z0Q
 
 | | Crystal Structure of OvoB | | Descriptor: | Aminotransferase, class I and II, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cai, Y.J, Huang, P, Wu, L, Zhou, J.H, Liu, P.H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | In Vitro Reconstitution of the Remaining Steps in Ovothiol A Biosynthesis: C-S Lyase and Methyltransferase Reactions.
Org. Lett., 20, 2018
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
9FZA
 
 | |
4N1U
 
 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
7TNC
 
 | | M13F/G116F Pseudomonas aeruginosa azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, CHLORIDE ION, ... | | Authors: | Liu, Y, Lu, Y. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for the Effects of Phenylalanine on Tuning the Reduction Potential of Type 1 Copper in Azurin.
Inorg.Chem., 62, 2023
|
|
7U2F
 
 | | G116F Pseudomonas aeruginosa azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Liu, Y, Lu, Y. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Effects of Phenylalanine on Tuning the Reduction Potential of Type 1 Copper in Azurin.
Inorg.Chem., 62, 2023
|
|
