7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7P9T
 
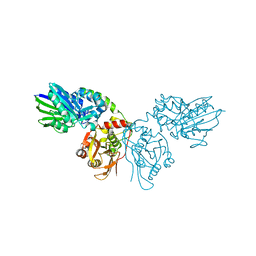 | | Crystal structure of CD73 in complex with dCMP in the open form | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PB5
 
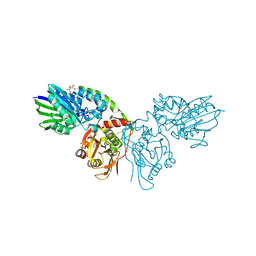 | | Crystal structure of CD73 in complex with UMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBA
 
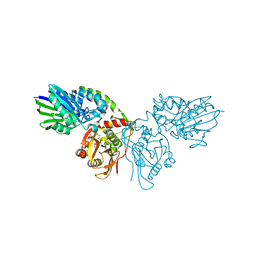 | | Crystal structure of CD73 in complex with IMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-08-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PD9
 
 | |
7PA4
 
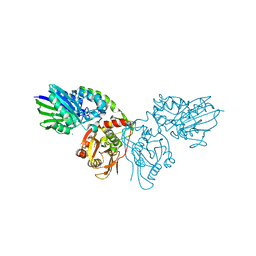 | | Crystal structure of CD73 in complex with CMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PBB
 
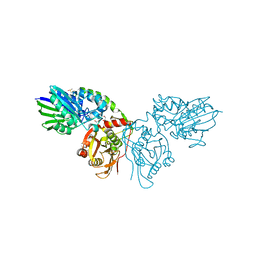 | |
7P9N
 
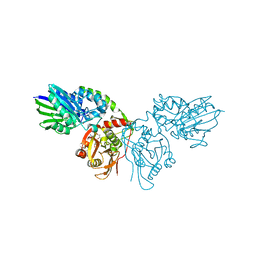 | | Crystal structure of CD73 in complex with AMP in the open form | | Descriptor: | 5'-nucleotidase, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7P9R
 
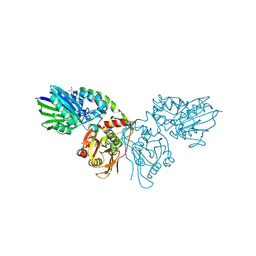 | | Crystal structure of CD73 in complex with GMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PCP
 
 | |
7PBY
 
 | |
7ZYT
 
 | | Crystal structure of the I318T pathogenic variant of the human dihydrolipoamide dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, ... | | Authors: | Nemes-Nikodem, E, Szabo, E, Vass, K.R, Lennartz, F, Nagy, B, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | Structural and Biochemical Investigation of Selected Pathogenic Mutants of the Human Dihydrolipoamide Dehydrogenase.
Int J Mol Sci, 24, 2023
|
|
8B4V
 
 | | X-ray structure of furin (PCSK3) in complex with benzamidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8B4W
 
 | | X-ray structure of furin (PCSK3) in complex with 1H-isoindol-3-amine | | Descriptor: | 1H-isoindol-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8B4X
 
 | | X-ray structure of furin (PCSK3) in complex with Guanidinomethyl-Phac-R-Tle-K-6-(aminomethyl)-3-amino-isoindol | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8CCI
 
 | | Crystal structure of Mycobacterium smegmatis thioredoxin reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fuesser, F.T, Koch, O, Kuemmel, D. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Novel starting points for fragment-based drug design against mycobacterial thioredoxin reductase identified using crystallographic fragment screening.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2ADW
 
 | |
1YSO
 
 | | YEAST CU, ZN SUPEROXIDE DISMUTASE WITH THE REDUCED BRIDGE BROKEN | | Descriptor: | COPPER (I) ION, YEAST CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Crane, B.R, Tsang, J, Tainer, J.A. | | Deposit date: | 1995-12-21 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
7NCX
 
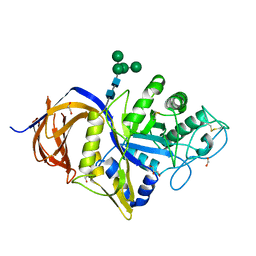 | | Crystal structure of GH30 (double mutant EE) from Thermothelomyces thermophila. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dimarogona, M, Nikolaivits, E, Topakas, E, Weiss, M, Feiler, C.G. | | Deposit date: | 2021-01-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Unique features of the bifunctional GH30 from Thermothelomyces thermophila revealed by structural and mutational studies
Carbohydrate Polymers, 273, 2021
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8OFO
 
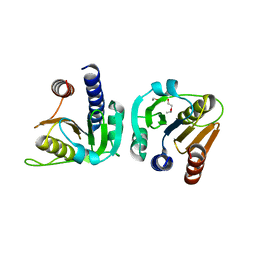 | | Structure of Enterococcus faecium CdaA | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Diadenylate cyclase | | Authors: | Garbers, T.B, Neumann, P, Ficner, R. | | Deposit date: | 2023-03-16 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8OFH
 
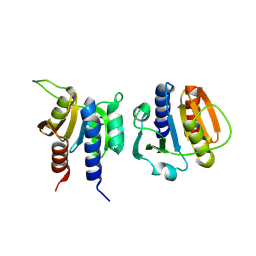 | | Streptococcus pneumoniae CdaA | | Descriptor: | Diadenylate cyclase | | Authors: | Garbers, T.B, Neumann, P, Ficner, R. | | Deposit date: | 2023-03-15 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8Q4S
 
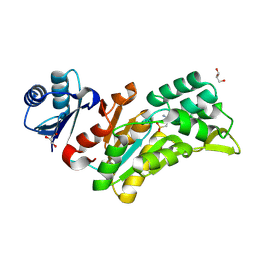 | |
8QOB
 
 | |
