3MFN
 
 | | Dfer_2879 protein of unknown function from Dyadobacter fermentans | | Descriptor: | ACETATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Chin, S, Eisen, J, Wu, D, Kerfeld, C, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-02 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | X-ray crystal structure of Dfer_2879 protein of unknown function from Dyadobacter fermentans.
To be Published
|
|
3B73
 
 | |
3ILK
 
 | | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structure of a probable methylase family protein from Haemophilus influenzae Rd KW20
To be Published
|
|
3C9P
 
 | | Crystal structure of uncharacterized protein SP1917 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein SP1917 | | Authors: | Chang, C, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of uncharacterized protein SP1917.
To be Published
|
|
4RA7
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-2-hydroxy-1-{[(2-hydroxynaphthalen-1-yl)carbonyl]amino}ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with nafcillin
To be Published
|
|
3GBY
 
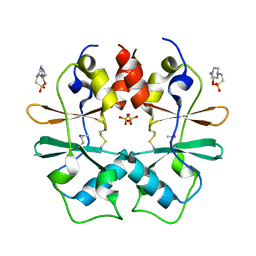 | | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Uncharacterized protein CT1051 | | Authors: | Fan, Y, Chang, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum
To be Published
|
|
1NNI
 
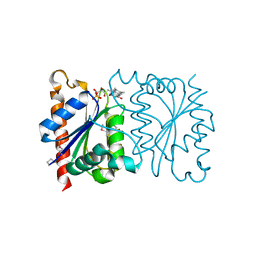 | | Azobenzene Reductase from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein yhda | | Authors: | Cuff, M.E, Kim, Y, Maj, L, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-13 | | Release date: | 2003-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azobenzene Reductase from Bacillus subtilis
To be Published, 2003
|
|
3G7R
 
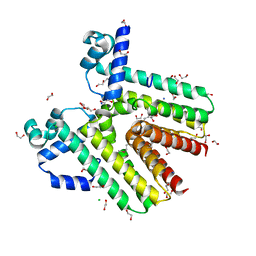 | | Crystal structure of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-10 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure and ligand specificity of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
4RTF
 
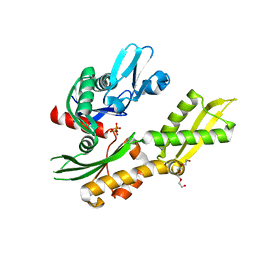 | | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv
To be Published
|
|
3M1R
 
 | | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Trevino, D, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-05 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The crystal structure of formimidoylglutamase from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
3M4R
 
 | | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-11 | | Release date: | 2010-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum
TO BE PUBLISHED
|
|
3GL5
 
 | | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, Putative DsbA oxidoreductase SCO1869, SODIUM ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor
To be Published
|
|
3MGL
 
 | |
3H35
 
 | | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2. | | Descriptor: | 1,2-ETHANEDIOL, S,R MESO-TARTARIC ACID, uncharacterized protein ABO_0056 | | Authors: | Cuff, M.E, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2.
TO BE PUBLISHED
|
|
3N04
 
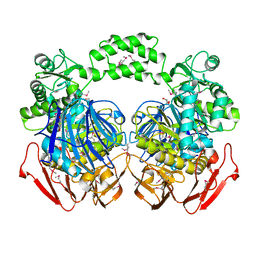 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 | | Descriptor: | GLYCEROL, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Freeman, L, Wilton, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (FAMILY 31) FROM RUMINOCOCCUS OBEUM ATCC 29174
Faseb J., 24, 2010
|
|
5BMZ
 
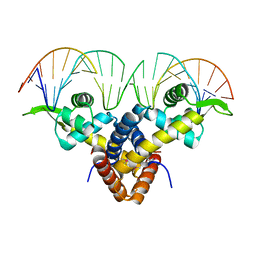 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA. | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*AP*TP*CP*AP*GP*TP*TP*AP*AP*AP*CP*TP*GP*AP*TP*AP*TP*TP*C)-3'), HcaR protein | | Authors: | Kim, Y, Joachimiak, G, Biglow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA.
To Be Published
|
|
4RYE
 
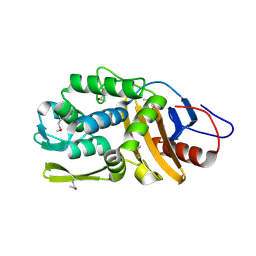 | | The crystal structure of D-ALANYL-D-ALANINE CARBOXYPEPTIDASE from Mycobacterium tuberculosis H37Rv | | Descriptor: | D-alanyl-D-alanine carboxypeptidase | | Authors: | Cuff, M, Tan, K, Hatzos-Skintges, C, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-12-15 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | The crystal structure of D-ALANYL-D-ALANINE CARBOXYPEPTIDASE from Mycobacterium tuberculosis H37Rv
To be Published
|
|
3H9P
 
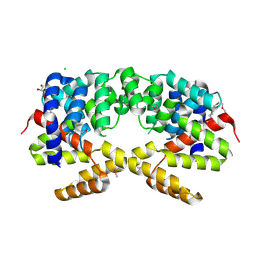 | | Crystal structure of putative triphosphoribosyl-dephospho-coA synthase from Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chang, C, Wu, R, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative triphosphoribosyl-dephospho-coA synthase from Archaeoglobus fulgidus
To be Published
|
|
3MR0
 
 | | Crystal Structure of Sensory Box Histidine Kinase/Response Regulator from Burkholderia thailandensis E264 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Tesar, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | Crystal Structure of Sensory Box Histidine Kinase/Response Regulator from Burkholderia thailandensis E264
To be Published
|
|
3MT0
 
 | | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1 | | Descriptor: | CHLORIDE ION, uncharacterized protein PA1789 | | Authors: | Tan, K, Chang, C, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | The crystal structure of a functionally unknown protein PA1789 from Pseudomonas aeruginosa PAO1
To be Published
|
|
3BRJ
 
 | | Crystal structure of mannitol operon repressor (MtlR) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mannitol operon repressor | | Authors: | Tan, K, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
1N2X
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | Authors: | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
3NKL
 
 | | Crystal Structure of UDP-D-Quinovosamine 4-Dehydrogenase from Vibrio fischeri | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Mack, J, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-20 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of UDP-D-Quinovosamine 4-Dehydrogenase from Vibrio fischeri
To be Published, 2010
|
|
3GVE
 
 | | Crystal structure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis | | Descriptor: | CITRIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Kim, Y, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Calcineurin-like Phosphoesterase from Bacillus subtilis
To be Published
|
|
3GK6
 
 | | Crystal structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS2. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Integron cassette protein VCH_CASS2 | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Integron Gene Cassette-Associated Protein from Vibrio cholerae Identifies a Cationic Drug-Binding Module.
Plos One, 6, 2011
|
|
