8W4O
 
 | | Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
4HSU
 
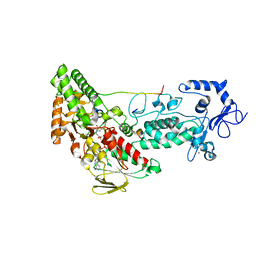 | | Crystal structure of LSD2-NPAC with H3(1-26)in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Xu, Y. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b.
Cell Res., 23, 2013
|
|
8W7A
 
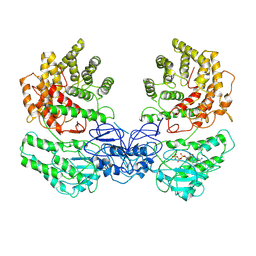 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC in the presence of GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Protein kinase domain-containing protein | | Authors: | Li, Y, Luo, M, Shao, K, Li, J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8WGO
 
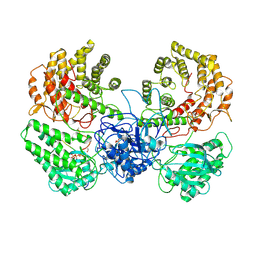 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC in the presence of PneA and GTPrS. | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-08-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8ZY2
 
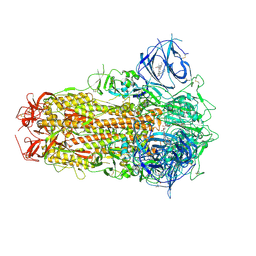 | | Sarbecovirus BANAL-20-52 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY9
 
 | | Ra9479 Bat ACE2 Dimer in Complex with Two BtKY72 Sarbecovirus Spike RBDs. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY7
 
 | | Sarbecovirus HeB2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY4
 
 | | Sarbecovirus YN2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY6
 
 | | Sarbecovirus GX2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY3
 
 | | Sarbecovirus BANAL-20-236 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY1
 
 | | Sarbecovirus BM48-31 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZYA
 
 | | Ra9479 Bat ACE2 Bound to BtkY72 Sarbecovirus Spike RBD (Focused Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY5
 
 | | Sarbecovirus RmYN02 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY0
 
 | | Sarbecovirus BtKY72 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
7KXE
 
 | |
7KXF
 
 | |
5XMM
 
 | | FLA-E*01801-167W/S | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
7MWN
 
 | | An engineered PYL2-based WIN 55,212-2 synthetic cannabinoid sensor with a stabilized HAB1 variant | | Descriptor: | ACETATE ION, Abscisic acid receptor PYL2, CHLORIDE ION, ... | | Authors: | Peterson, F.C, Beltran, J, Bedewitz, M, Steiner, P.J, Cutler, S.R, Whitehead, T.A. | | Deposit date: | 2021-05-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Rapid biosensor development using plant hormone receptors as reprogrammable scaffolds.
Nat.Biotechnol., 40, 2022
|
|
4LAL
 
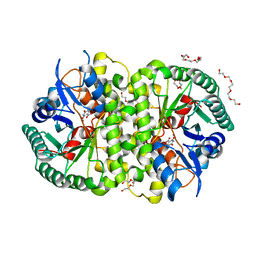 | | Crystal structure of Cordyceps militaris IDCase D323A mutant in complex with 5-carboxyl-uracil | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAM
 
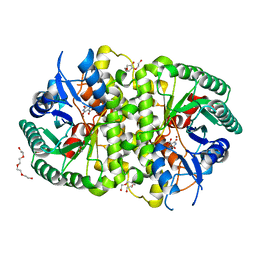 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in complex with 5-carboxyl-uracil | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, HEXAETHYLENE GLYCOL, Uracil-5-carboxylate decarboxylase, ... | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAN
 
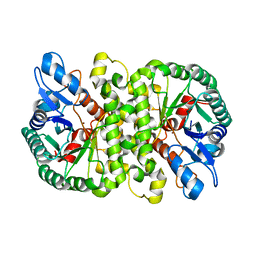 | | Crystal structure of Cordyceps militaris IDCase H195A mutant | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4LAO
 
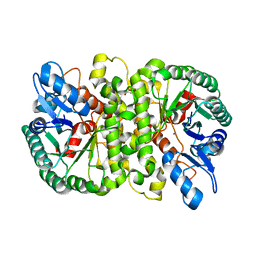 | | Crystal structure of Cordyceps militaris IDCase H195A mutant (Zn) | | Descriptor: | Cordyceps militaris IDCase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
8F6R
 
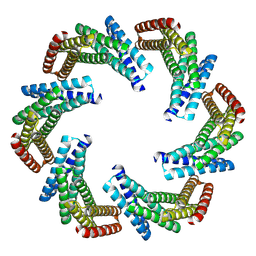 | | CryoEM structure of designed modular protein oligomer C6-79 | | Descriptor: | De novo designed oligomeric protein C6-79 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
8F6Q
 
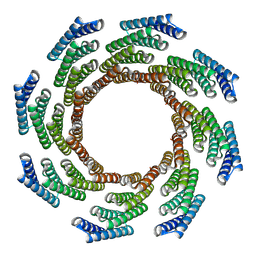 | | CryoEM structure of designed modular protein oligomer C8-71 | | Descriptor: | C8-71 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2022-11-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies.
Cell, 187, 2024
|
|
