3CGH
 
 | |
8IUA
 
 | |
8IU8
 
 | |
8IU9
 
 | |
8IUB
 
 | |
8IUC
 
 | |
2F46
 
 | |
2JVA
 
 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | Descriptor: | Peptidyl-tRNA hydrolase domain protein | | Authors: | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
3K5J
 
 | |
3HSA
 
 | |
3KK7
 
 | |
3F1Z
 
 | |
3NNQ
 
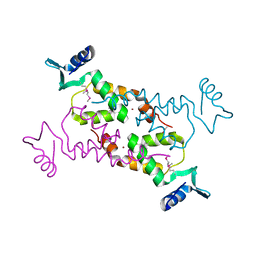 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | ACETATE ION, N-terminal domain of Moloney murine leukemia virus integrase, ZINC ION | | Authors: | Guan, R, Xiao, R, Acton, T, Jiang, M, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-14 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
3DEE
 
 | |
5JJG
 
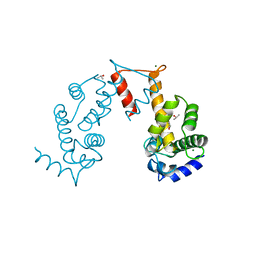 | | Structure of magnesium-loaded ALG-2 | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Pcalcium-binding protein ALG-2, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2016-04-23 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | EF5 Is the High-Affinity Mg(2+) Site in ALG-2.
Biochemistry, 55, 2016
|
|
3OGI
 
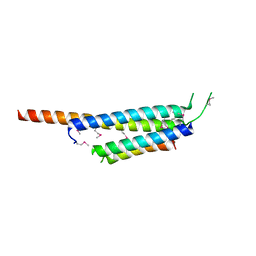 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP complex (Rv2346c-Rv2347c) | | Descriptor: | Putative ESAT-6-like protein 6, Putative ESAT-6-like protein 7 | | Authors: | Arbing, M.A, Chan, S, Zhou, T.T, Ahn, C, Harris, L, Kuo, E, Sawaya, M.R, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-16 | | Release date: | 2010-08-25 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3GF8
 
 | |
3DUE
 
 | |
3DCX
 
 | |
3PGE
 
 | | Structure of sumoylated PCNA | | Descriptor: | Proliferating cell nuclear antigen, SUMO-modified proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Brogie, J.E, Gakhar, L, Washington, T. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of SUMO-Modified Proliferating Cell Nuclear Antigen.
J.Mol.Biol., 406, 2011
|
|
2FEA
 
 | |
7M42
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of neutralizing antibodies REGN10985 and REGN10989 | | Descriptor: | REGN10985 antibody Fab fragment heavy chain, REGN10985 antibody Fab fragment light chain, REGN10989 antibody Fab fragment heavy chain, ... | | Authors: | Zhou, Y, Romero Hernandez, A, Saotome, K, Franklin, M.C. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies.
Cell, 184, 2021
|
|
3HBZ
 
 | |
3H41
 
 | |
2FNO
 
 | |
