6BID
 
 | | 1.15 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(8S,11S,14S)-11-(cyclohexylmethyl)-8-(hydroxymethyl)-5,10,13-trioxo-1,4,9,12,17,18-hexaazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6B3E
 
 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
6B0X
 
 | |
6DHC
 
 | | X-ray structure of BACE1 in complex with a bicyclic isoxazoline carboxamide as the P3 ligand | | Descriptor: | (3R,3aR,6aS)-N-[(4R,7S,8S,10R,13S)-8-hydroxy-10,17-dimethyl-7-(2-methylpropyl)-5,11,14-trioxo-13-(propan-2-yl)-2-thia-6,12,15-triazaoctadecan-4-yl]hexahydrofuro[3,2-d][1,2]oxazole-3-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Mesecar, A.D, Lendy, E.K. | | Deposit date: | 2018-05-19 | | Release date: | 2018-07-25 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design, synthesis, X-ray studies, and biological evaluation of novel BACE1 inhibitors with bicyclic isoxazoline carboxamides as the P3 ligand.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6B23
 
 | |
6C21
 
 | |
6BWK
 
 | |
6DLU
 
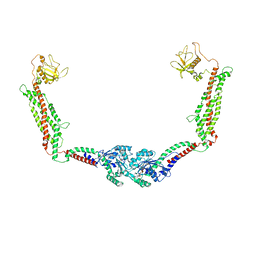 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | Descriptor: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | Deposit date: | 2018-06-02 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
1A5Y
 
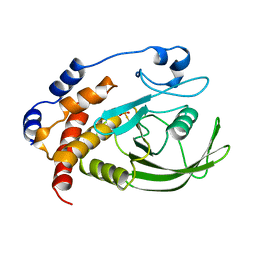 | |
3ZO7
 
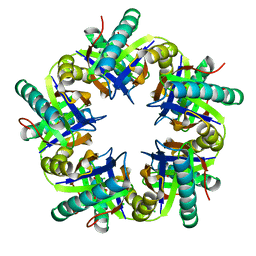 | | Crystal structure of ClcFE27A with substrate | | Descriptor: | (2S)-2-chloranyl-2-[(2R)-5-oxidanylidene-2H-furan-2-yl]ethanoic acid, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
7MSF
 
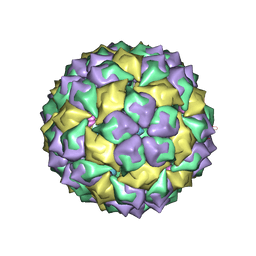 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*AP*AP*CP*AP*GP*GP*CP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-20 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
7JTJ
 
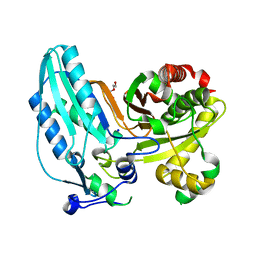 | |
7KUA
 
 | | Crystal structure of the MarR family transcriptional regulator from Pseudomonas putida bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Transcriptional regulator, MarR family | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7L19
 
 | | Crystal structure of the MarR family transcriptional regulator from Enterobacter soli strain LF7 bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family transcriptional regulator, NICKEL (II) ION | | Authors: | Lietzan, A.D, Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7L1I
 
 | | Crystal structure of the MarR family transcriptional regulator from Acineotobacter baumannii bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family multidrug resistance pump transcriptional regulator, NICKEL (II) ION | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7LKS
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2f | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKX
 
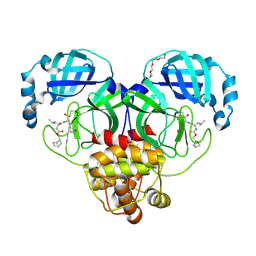 | | 1.60 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3e | | Descriptor: | (1R,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKT
 
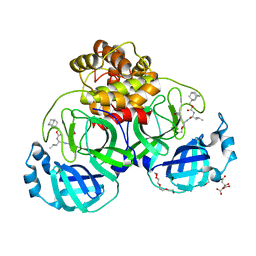 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | Descriptor: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKR
 
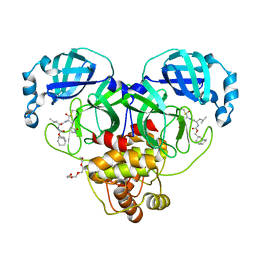 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2a | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKV
 
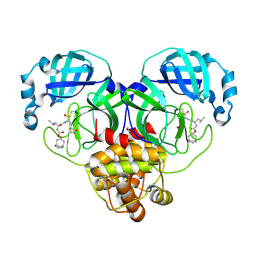 | | 1.55 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKW
 
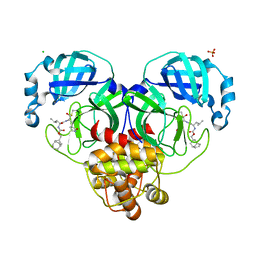 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3d (deuterated analog of inhibitor 3c) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKU
 
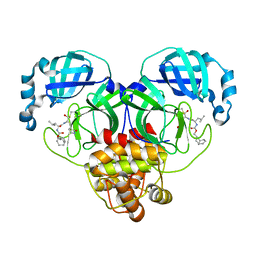 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3b (deuterated analog of inhibitor 2a) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LS2
 
 | | 80S ribosome from mouse bound to eEF2 (Class I) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
7LS1
 
 | | 80S ribosome from mouse bound to eEF2 (Class II) | | Descriptor: | 28S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
7L7V
 
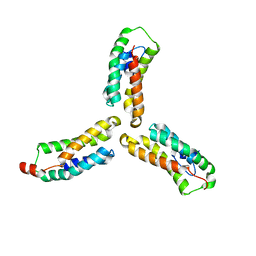 | | Crystal structure of Arabidopsis NRG1.1 CC-R domain K94E/K96E/R99E/K100E/R103E/K106E/K110E mutant | | Descriptor: | Probable disease resistance protein At5g66900 | | Authors: | Walton, W.G, Wan, L, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plant "helper" immune receptors are Ca 2+ -permeable nonselective cation channels.
Science, 373, 2021
|
|
