7EAX
 
 | | Crystal complex of p53-V272M and antimony ion | | Descriptor: | ANTIMONY (III) ION, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Tang, Y. | | Deposit date: | 2021-03-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Repurposing antiparasitic antimonials to noncovalently rescue temperature-sensitive p53 mutations.
Cell Rep, 39, 2022
|
|
7E5X
 
 | |
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7F0H
 
 | |
7JU8
 
 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
5YKR
 
 | |
5YKT
 
 | |
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
8VX0
 
 | | CRYSTAL STRUCTURE OF CYP2C9*14 IN COMPLEX WITH LOSARTAN | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Shah, M.B. | | Deposit date: | 2024-02-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and biophysical analysis of cytochrome P450 2C9*14 and *27 variants in complex with losartan.
J.Inorg.Biochem., 258, 2024
|
|
8VZ7
 
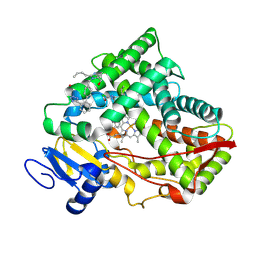 | |
9EOW
 
 | |
8Y1J
 
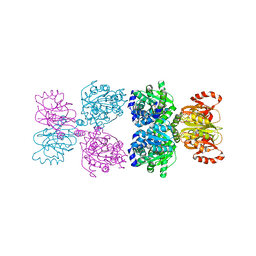 | |
8GRR
 
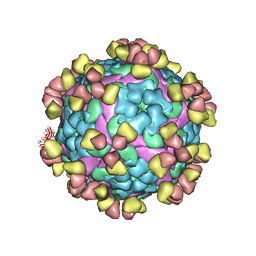 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W125 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Kun, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8GSP
 
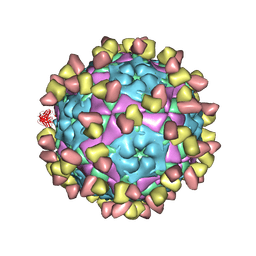 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W2 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
8HC0
 
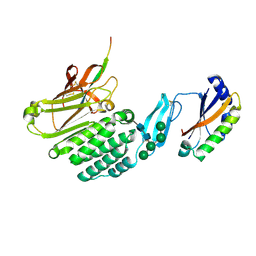 | | Crystal structure of the extracellular domains of GPR110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor F1, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, F.F, Song, G.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Extracellular Domains of GPR110.
J.Mol.Biol., 435, 2023
|
|
7WHD
 
 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Wang, X.W, Ge, J.W, Wang, Z.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
7VK9
 
 | | Crystal structure of xCas9 P411T | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | Bao, R, Liu, H.Y, Luo, Y.Z, Song, Y.J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Acs Catalysis, 12, 2022
|
|
7VAH
 
 | |
8I8F
 
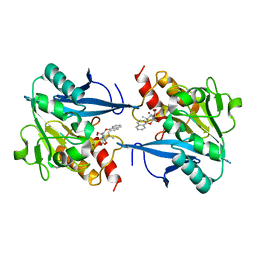 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed compound 1 | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-(2-naphthalen-1-yloxyethanoylamino)-2-oxidanyl-2-oxidanylidene-ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Shi, X, Liu, W. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
7WH8
 
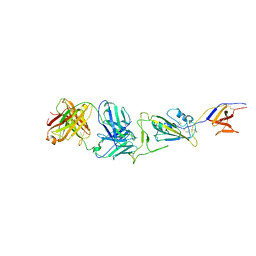 | |
7WHB
 
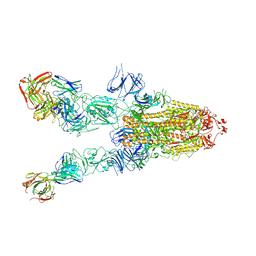 | | SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zeng, J.W, Ge, J.W, Wang, X.Q. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters 1 .
Emerg Microbes Infect, 12, 2023
|
|
