6JCJ
 
 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5XSJ
 
 | | XylFII-LytSN complex | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS, ... | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
6IR5
 
 | | P domain of GII.3-TV24 | | Descriptor: | VP1 Capsid protein | | Authors: | Yang, Y. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IS5
 
 | | P domain of GII.3-TV24 with A-tetrasaccharide complex | | Descriptor: | VP1 Capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yang, Y. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
7V9U
 
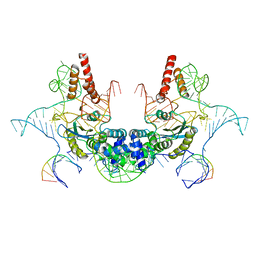 | | Cryo-EM structure of E.coli retron-Ec86 (RT-msDNA-RNA) at 3.2 angstrom | | Descriptor: | DNA (105-MER), RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*G)-3'), RNA (81-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
9KBG
 
 | | The structure of B19V NS1_2-570/AMPPNP | | Descriptor: | MAGNESIUM ION, Non-structural protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gan, J, Zhang, Y. | | Deposit date: | 2024-10-30 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural and functional studies of the main replication protein NS1 of human parvovirus B19.
Nucleic Acids Res., 53, 2025
|
|
7C53
 
 | |
9KBJ
 
 | | The structure of B19V NS1_200-501/AMPPNP | | Descriptor: | MAGNESIUM ION, Non-structural protein 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gan, J, Zhang, Y. | | Deposit date: | 2024-10-30 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional studies of the main replication protein NS1 of human parvovirus B19.
Nucleic Acids Res., 53, 2025
|
|
9KBH
 
 | | The structure of B19V NS1_2-570/ssDNA/AMPPNP | | Descriptor: | DNA (67-MER), MAGNESIUM ION, Non-structural protein 1, ... | | Authors: | Gan, J, Zhang, Y. | | Deposit date: | 2024-10-30 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional studies of the main replication protein NS1 of human parvovirus B19.
Nucleic Acids Res., 53, 2025
|
|
9KBI
 
 | | The structure of B19V NS1_2-570/dsDNA/AMPPNP | | Descriptor: | DNA (48-MER), MAGNESIUM ION, Non-structural protein 1, ... | | Authors: | Gan, J, Zhang, Y. | | Deposit date: | 2024-10-30 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structural and functional studies of the main replication protein NS1 of human parvovirus B19.
Nucleic Acids Res., 53, 2025
|
|
8GWA
 
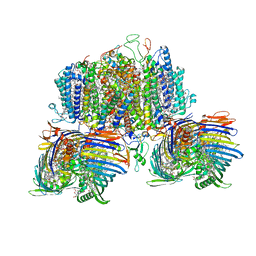 | | Structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-electron microscopy structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium.
J Integr Plant Biol, 65, 2023
|
|
9HBO
 
 | |
9D52
 
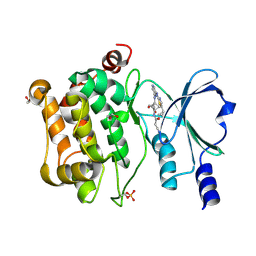 | | Structure of PAK4 in complex with compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-N-[3-({6-[(5-cyclopropyl-1,3-thiazol-2-yl)amino]pyrazin-2-yl}amino)bicyclo[1.1.1]pentan-1-yl]acetamide, Serine/threonine-protein kinase PAK 4 | | Authors: | Boone, C, Suto, R, Olland, A. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
8H1R
 
 | | Crystal structure of LptDE-YifL complex | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Surface lipoprotein sorting by crosstalk between Lpt and Lol pathways in gram-negative bacteria
Nat Commun, 16, 2025
|
|
8H1S
 
 | | Crystal structure of apo-LptDE complex | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Surface lipoprotein sorting by crosstalk between Lpt and Lol pathways in gram-negative bacteria
Nat Commun, 16, 2025
|
|
5MIO
 
 | | KIF2C-DARPIN FUSION PROTEIN BOUND TO TUBULIN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF2C,KIF2C FUSED TO A DARPIN,KIF2C FUSED TO A DARPIN, ... | | Authors: | Wang, W, Gigant, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin.
Nat Commun, 8, 2017
|
|
8GQD
 
 | | Complex Structure of Arginine Kinase McsB and McsA from Staphylococcus aureus | | Descriptor: | Protein-arginine kinase, Protein-arginine kinase activator protein, ZINC ION | | Authors: | Lu, K, Luo, B, Tao, X, Li, H, Xie, Y, Zhao, Z, Xia, W, Su, Z, Mao, Z. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Complex structure and activation mechanism of arginine kinase McsB by McsA.
Nat.Chem.Biol., 2024
|
|
1I37
 
 | |
1I38
 
 | |
8I8Y
 
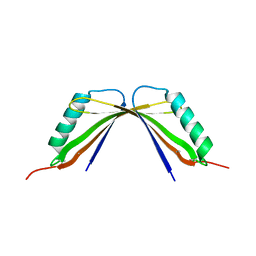 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6J0Q
 
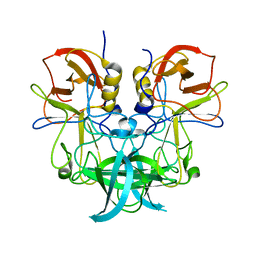 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
