7YVK
 
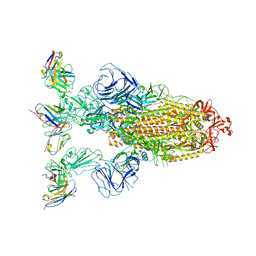 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH272 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH272 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVO
 
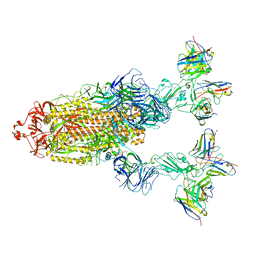 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH027/132 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH132 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVH
 
 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH132 Fab | | Descriptor: | Spike glycoprotein, TH132 Fab heavy chain, TH132 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVM
 
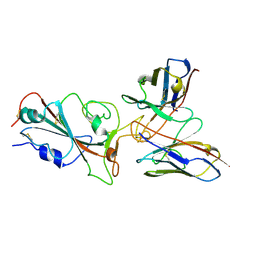 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH272 Fab | | Descriptor: | Spike glycoprotein, TH281 Fab heavy chain, TH281 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVF
 
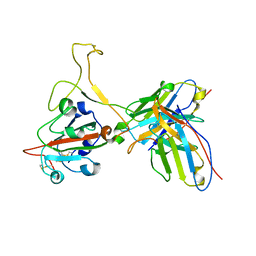 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH027 Fab | | Descriptor: | Spike glycoprotein, TH027 Fab heavy chain, TH027 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVJ
 
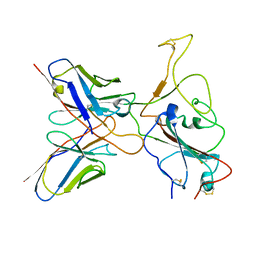 | | Omicron BA.4/5 SARS-CoV-2 S RBD in complex with TH236 Fab | | Descriptor: | Spike glycoprotein, TH236 Fab heavy chain, TH236 Fab light chain | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7YVI
 
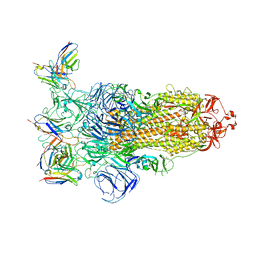 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH236 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
6U2H
 
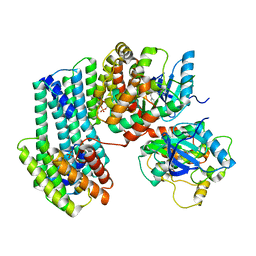 | | BRAF dimer bound to 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8Z9C
 
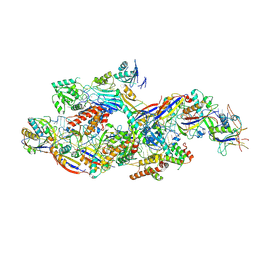 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | Protein structure, RNA (41-MER), RNA (48-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHE
 
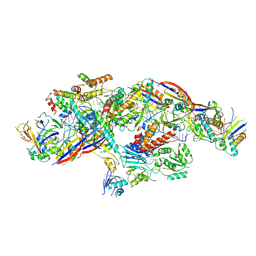 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state II | | Descriptor: | RNA (29-MER), RNA (46-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4J
 
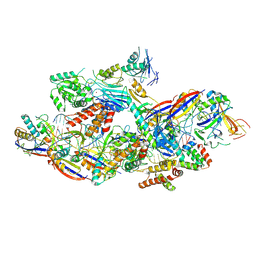 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (38-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4L
 
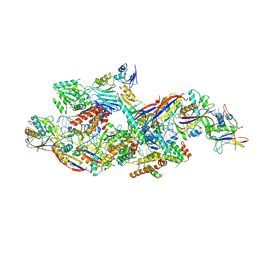 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z9E
 
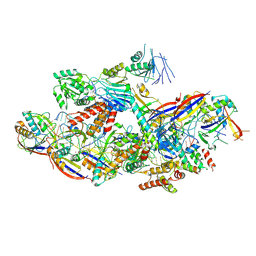 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (39-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z99
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I | | Descriptor: | RNA (49-MER), RNA (54-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHD
 
 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state I | | Descriptor: | RNA (35-MER), RNA (53-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8QV5
 
 | | Structure of human SPNS2 in LMNG | | Descriptor: | Nanobody D12, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Li, H.Z, Pike, A.C.W, McKinley, G, Mukhopadhyay, S.M.M, Moreau, C, Scacioc, A, Abrusci, P, Borkowska, O, Chalk, R, Stefanic, S, Burgess-Brown, N, Duerr, K.L, Sauer, D.B. | | Deposit date: | 2023-10-17 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Transport and inhibition of the sphingosine-1-phosphate exporter SPNS2.
Nat Commun, 16, 2025
|
|
8QV6
 
 | | Structure of human SPNS2 in DDM | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody D12, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Li, H.Z, Pike, A.C.W, McKinley, G, Mukhopadhyay, S.M.M, Moreau, C, Scacioc, A, Abrusci, P, Borkowska, O, Chalk, R, Stefanic, S, Burgess-Brown, N, Duerr, K.L, Sauer, D.B. | | Deposit date: | 2023-10-17 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Transport and inhibition of the sphingosine-1-phosphate exporter SPNS2.
Nat Commun, 16, 2025
|
|
5V3B
 
 | |
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
2DKV
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, chitinase | | Authors: | Kezuka, Y, Nishizawa, Y, Watanabe, T, Nonaka, T. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
8CLN
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus, SeMet derivative for SAD phasing | | Descriptor: | Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLO
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
3OCX
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase mutant D66N complexed with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
7RWF
 
 | | Crystal structure of CDK2 in complex with TW8672 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
3OCW
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase mutant D66N complexed with 3'-AMP | | Descriptor: | Lipoprotein E, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl] dihydrogen phosphate | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
