7ROY
 
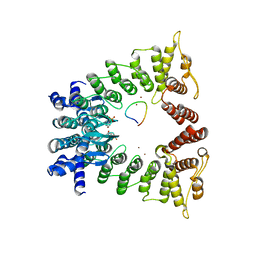 | | The structure of the Fem1B:FNIP1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Folliculin-interacting protein 1, Protein fem-1 homolog B, ... | | Authors: | Gee, C.L, Mena, E.L, Manford, A.G, Rape, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis and regulation of the reductive stress response.
Cell, 184, 2021
|
|
6QW5
 
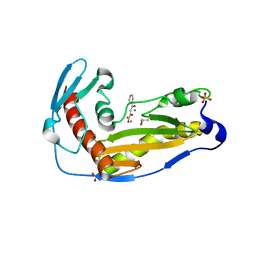 | | Structure and function of the toscana virus cap snatching endonuclease | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, RNA-dependent RNA polymerase, ... | | Authors: | Reguera, J, Jones, R, Bragagniolo, G, Lessoued, S, Mate, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-25 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structure and function of the Toscana virus cap-snatching endonuclease.
Nucleic Acids Res., 47, 2019
|
|
6QVV
 
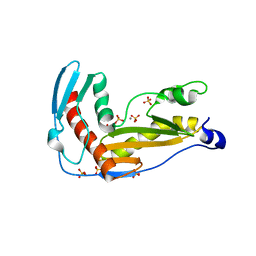 | | Structure and function of phenuiviridae cap snatching endonucleases | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA-dependent RNA polymerase, ... | | Authors: | Reguera, J, Jones, R, Bragagniolo, G, Lessoued, S, Mate, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of the Toscana virus cap-snatching endonuclease.
Nucleic Acids Res., 47, 2019
|
|
6QW0
 
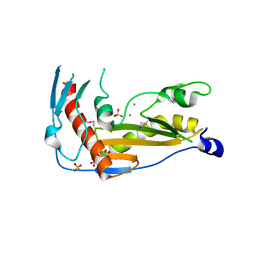 | | Structure and function of toscana virus cap snatching endonucleases | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA-dependent RNA polymerase, ... | | Authors: | Reguera, J, Jones, R, Bragagniolo, G, Lessoued, S, Mate, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the Toscana virus cap-snatching endonuclease.
Nucleic Acids Res., 47, 2019
|
|
6GSI
 
 | |
6GSH
 
 | | Feline Calicivirus Strain F9 | | Descriptor: | POTASSIUM ION, VP1 | | Authors: | Conley, M.J, Bhella, D. | | Deposit date: | 2018-06-14 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Calicivirus VP2 forms a portal-like assembly following receptor engagement.
Nature, 565, 2019
|
|
1XIN
 
 | |
7BLV
 
 | |
7BM0
 
 | | Crystal structure of the tick-borne encephalitis virus NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase domain, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anindita, P.D, Grinkevich, P, Franta, Z. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
5GTU
 
 | |
6B9Q
 
 | |
7SPT
 
 | | Crystal structure of exofacial state human glucose transporter GLUT3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
3CHO
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with 2-amino-N-[4-(phenylmethoxy)phenyl]-acetamide | | Descriptor: | ACETATE ION, Leukotriene A-4 hydrolase, N-[4-(benzyloxy)phenyl]glycinamide, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
7SPS
 
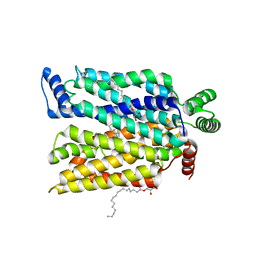 | | Crystal structure of human glucose transporter GLUT3 bound with exofacial inhibitor SA47 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
6CRG
 
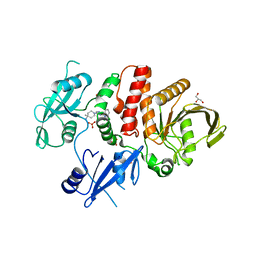 | | Crystal Structure of Shp2 E76K GOF Mutant in complex with SHP099 | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2018-03-17 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition.
Nat Commun, 9, 2018
|
|
3CHS
 
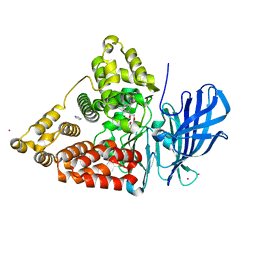 | | Crystal structure of leukotriene A4 hydrolase in complex with (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid | | Descriptor: | (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CHP
 
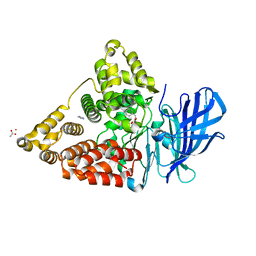 | | Crystal structure of leukotriene a4 hydrolase in complex with (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid | | Descriptor: | (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
6CRF
 
 | | Crystal Structure of Shp2 E76K GOF Mutant in the Open Conformation | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2018-03-17 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition.
Nat Commun, 9, 2018
|
|
6BN5
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 F285S in Complex with Allosteric Inhibitor JLR-2 | | Descriptor: | 3-benzyl-8-chloro-2-hydroxy-4H-pyrimido[2,1-b][1,3]benzothiazol-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Blacklow, S.C, Stams, T, Fodor, M, LaRochelle, J.R. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Identification of an allosteric benzothiazolopyrimidone inhibitor of the oncogenic protein tyrosine phosphatase SHP2.
Bioorg. Med. Chem., 25, 2017
|
|
7U8Z
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with fluorinated peptoid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)benzamido}methyl)-3-fluoro-N-hydroxybenzamide, ACETATE ION, ... | | Authors: | Watson, P.R, Cragin, A.D, Christianson, D.W. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Fluorinated Peptoid-Based Histone Deacetylase (HDAC) Inhibitors for Therapy-Resistant Acute Leukemia.
J.Med.Chem., 65, 2022
|
|
3LSA
 
 | | Padron0.9-OFF (non-fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
7U4P
 
 | |
3LS3
 
 | | Padron0.9-ON (fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
6PYE
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NR160 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-[(1-benzyl-1H-tetrazol-5-yl)methyl]-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-2-(trifluoromethyl)benzamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-07-29 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.480003 Å) | | Cite: | Multicomponent Synthesis, Binding Mode, and Structure-Activity Relationship of Selective Histone Deacetylase 6 (HDAC6) Inhibitors with Bifurcated Capping Groups.
J.Med.Chem., 63, 2020
|
|
6CEL
 
 | | CBH1 (E212Q) CELLOPENTAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
