4MYO
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
4PV4
 
 | | Proline aminopeptidase P II from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-14 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Proline aminopeptidase P II from Yersinia pestis
To be Published
|
|
4MWA
 
 | | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Brunzelle, J.S, Xu, X, Cui, H, Maltseva, N, Bishop, B, Kwon, K, Savchenko, A, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of GCPE Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4MY9
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MALONATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5893 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor C91
To be Published
|
|
4MVA
 
 | | 1.43 Angstrom Resolution Crystal Structure of Triosephosphate Isomerase (tpiA) from Escherichia coli in Complex with Acetyl Phosphate. | | Descriptor: | 1,2-ETHANEDIOL, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M.L, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
4N1X
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G | | Descriptor: | OPEN FORM - PENICILLIN G, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-10-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G
To be Published
|
|
4O4A
 
 | | 2.75 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis. | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Lipoprotein, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4ODI
 
 | | 2.6 Angstrom Crystal Structure of Putative Phosphoglycerate Mutase 1 from Toxoplasma gondii | | Descriptor: | Phosphoglycerate mutase PGMII, SODIUM ION | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-10 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4OHN
 
 | | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, HISTIDINE | | Authors: | Brunzelle, J.S, Wawrzak, W, Yim, Y, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of an ABC uptake transporter substrate binding protein from Streptococcus pneumoniae with Bound Histidine
To be Published
|
|
4OMV
 
 | | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4 | | Descriptor: | Macrophage growth locus, subunit A | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-27 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Putative Macrophage Growth Locus, subunit A From Francisella tularensis SCHU S4
TO BE PUBLISHED
|
|
4MYX
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-5-{[(2-{3-[(1E)-N-hydroxyethanimidoyl]phenyl}propan-2-yl)carbamoyl]amino}benzamide, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Bacillus anthracis str. Ame complexed with P32
To be Published
|
|
4MYA
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110 | | Descriptor: | 4-{(1R)-1-[1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl]ethoxy}quinolin-2(1H)-one, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8997 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor A110
To be Published
|
|
4MY8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Kavitha, M, Cuny, G, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2924 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21
To be Published, 2013
|
|
4MZ8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with an Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-29 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5004 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound C91
To be Published
|
|
4MZ1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 | | Descriptor: | 1-(4-bromophenyl)-3-{2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}urea, ACETIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3991 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12
To be Published, 2013
|
|
4MQB
 
 | | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
To be Published
|
|
4MPH
 
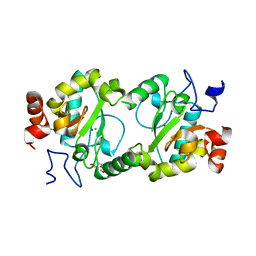 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-bound | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0301 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
4PUP
 
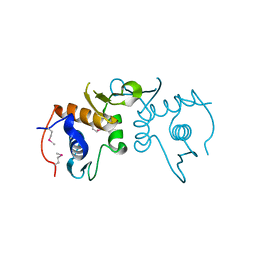 | | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Burkholderia cenocepacia J2315 | | Descriptor: | Uncharacterized protein | | Authors: | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Minasov, G, Jedrzejczak, R, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-13 | | Release date: | 2014-04-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Burkholderia cenocepacia J2315
To be Published
|
|
4PWZ
 
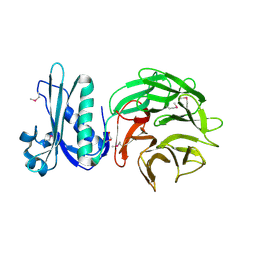 | | Crystal structure of TolB protein from Yersinia pestis CO92 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal structure of TolB protein from Yersinia pestis CO92
To be Published
|
|
4PZL
 
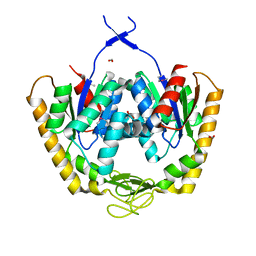 | | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Adenylate kinase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4QNE
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with NAD and IMP | | Descriptor: | INOSINIC ACID, Inosine 5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with NAD and IMP
To be Published
|
|
4Q32
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(naphthalen-2-yl)-2-[2-(pyridin-2-yl)-1H-benzimidazol-1-yl]acetamide | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and C91
To be Published
|
|
4PZ0
 
 | | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2) | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2).
To be Published
|
|
4PWT
 
 | | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from Yersinia pestis CO92 | | Descriptor: | FORMIC ACID, PYROPHOSPHATE 2-, Peptidoglycan-associated lipoprotein, ... | | Authors: | Maltseva, N, Kim, Y, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal structure of peptidoglycan-associated outer membrane lipoprotein from
Yersinia pestis CO92
To be Published
|
|
4PZJ
 
 | | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243
To be Published
|
|
