2L65
 
 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
6L69
 
 | | Crystal structure of CYP154C2 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
6LOI
 
 | |
7DGB
 
 | |
7DHJ
 
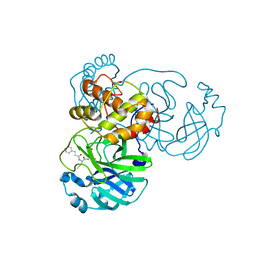 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
7DGH
 
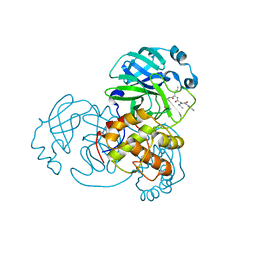 | |
7DGF
 
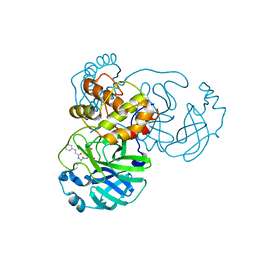 | |
7DGG
 
 | |
7DGI
 
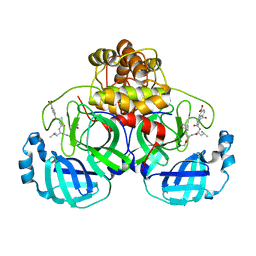 | |
2L6H
 
 | |
2L6F
 
 | | NMR Solution structure of FAT domain of FAK complexed with LD2 and LD4 motifs of PAXILLIN | | Descriptor: | Focal adhesion kinase 1, linker1, Paxillin, ... | | Authors: | Bertolucci, C.M, Guibao, C, Zhang, C, Zheng, J. | | Deposit date: | 2010-11-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Fat Domain of Fak Complexed with Ld2 and Ld4 Motifs of Paxillin
To be Published
|
|
7VUZ
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with PAMP-12, state2 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV4
 
 | |
7VDM
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with substance P | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV5
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state1 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VDL
 
 | |
7VDH
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state2 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV0
 
 | |
7VV6
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Mas-related G-protein coupled receptor member X2 | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VUY
 
 | |
7VV3
 
 | |
7W92
 
 | | Open state of SARS-CoV-2 Delta variant spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9E
 
 | | SARS-CoV-2 Delta S-8D3 | | Descriptor: | Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain, Spike glycoprotein, The light chain of 8D3 fab | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W99
 
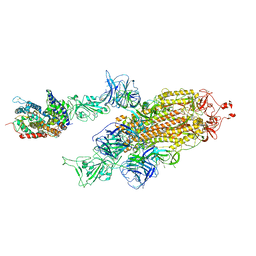 | | SARS-CoV-2 Delta S-ACE2-C2a | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W94
 
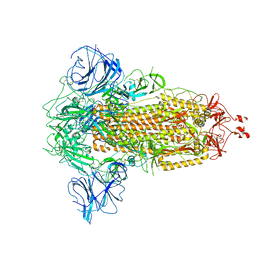 | |
