4J0E
 
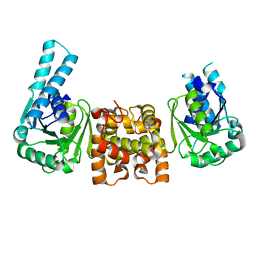 | |
4J0F
 
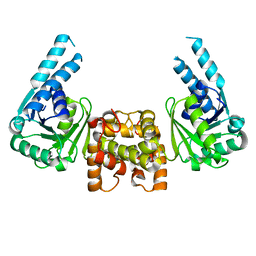 | |
3BOH
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 1 with acetate (CDCA1-R1) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3BOE
 
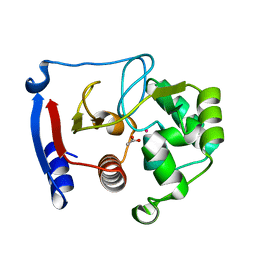 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 with acetate (CDCA1-R2) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
1U9I
 
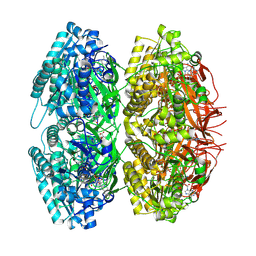 | | Crystal Structure of Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KaiC, MAGNESIUM ION | | Authors: | Xu, Y, Mori, T, Pattanayek, R, Pattanayek, S, Egli, M, Johnson, C.H. | | Deposit date: | 2004-08-09 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
3BOC
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- zinc bound domain 2 (CDCA1-R2) | | Descriptor: | Cadmium-specific carbonic anhydrase, ZINC ION | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3BOB
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 | | Descriptor: | CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
1UL3
 
 | | Crystal Structure of PII from Synechocystis sp. PCC 6803 | | Descriptor: | CALCIUM ION, GLYCEROL, Nitrogen regulatory protein P-II | | Authors: | Xu, Y, Carr, P.D, Clancy, P, Garcia-Dominguez, M, Forchhammer, K, Florencio, F, Tandeau de Marsac, N, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2003-09-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1V4A
 
 | | Structure of the N-terminal Domain of Escherichia coli Glutamine Synthetase adenylyltransferase | | Descriptor: | Glutamate-ammonia-ligase adenylyltransferase | | Authors: | Xu, Y, Zhang, R, Joachimiak, A, Carr, P.D, Ollis, D.L, Vasudevan, S.G. | | Deposit date: | 2003-11-12 | | Release date: | 2004-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the n-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase
Structure, 12, 2004
|
|
1NH5
 
 | | AUTOMATIC ASSIGNMENT OF NMR DATA AND DETERMINATION OF THE PROTEIN STRUCTURE OF A NEW WORLD SCORPION NEUROTOXIN USING NOAH/DIAMOD | | Descriptor: | Neurotoxin 5 | | Authors: | Xu, Y, Jablonsky, M.J, Jackson, P.L, Krishna, N.R, Braun, W. | | Deposit date: | 2002-12-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Automatic assignment of NOESY Cross peaks and determination of the protein structure of a new world scorpion neurotoxin Using NOAH/DIAMOD
J.Magn.Reson., 148, 2001
|
|
2H25
 
 | |
1QR6
 
 | | HUMAN MITOCHONDRIAL NAD(P)-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Xu, Y, Bhargava, G, Wu, H, Loeber, G, Tong, L. | | Deposit date: | 1999-06-18 | | Release date: | 1999-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human mitochondrial NAD(P)(+)-dependent malic enzyme: a new class of oxidative decarboxylases.
Structure, 7, 1999
|
|
1WDF
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
1WDG
 
 | | crystal structure of MHV spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Qin, L, Li, X, Bai, Z, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-05-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Basis for Coronavirus-mediated Membrane Fusion: CRYSTAL STRUCTURE OF MOUSE HEPATITIS VIRUS SPIKE PROTEIN FUSION CORE
J.Biol.Chem., 279, 2004
|
|
1WP7
 
 | | crystal structure of Nipah Virus fusion core | | Descriptor: | fusion protein | | Authors: | Xu, Y, Liu, Y, Lou, Z, Su, N, Bai, Z, Gao, G.F, Rao, Z. | | Deposit date: | 2004-08-31 | | Release date: | 2005-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nipah Virus fusion core
To be Published
|
|
1WNC
 
 | | Crystal structure of the SARS-CoV Spike protein fusion core | | Descriptor: | E2 glycoprotein | | Authors: | Xu, Y, Lou, Z, Liu, Y, Pang, H, Tien, P, Gao, G.F, Rao, Z. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core
J.Biol.Chem., 279, 2004
|
|
7S8V
 
 | |
7S0Q
 
 | |
7WKD
 
 | | TRH-TRHR G protein complex | | Descriptor: | Gq, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Y, Cai, H, You, C, He, X, Yuan, Q, Jiang, H, Cheng, X, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into ligand binding and activation of the human thyrotropin-releasing hormone receptor.
Cell Res., 32, 2022
|
|
7F6D
 
 | | Reconstruction of the HerA-NurA complex from Deinococcus radiodurans | | Descriptor: | HerA, NurA | | Authors: | Xu, Y, Xu, L, Guo, J, Hua, Y, Zhao, Y. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Mechanisms of helicase activated DNA end resection in bacteria.
Structure, 30, 2022
|
|
1BIK
 
 | | X-RAY STRUCTURE OF BIKUNIN FROM THE HUMAN INTER-ALPHA-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIKUNIN, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Guss, J.M, Ollis, D.L. | | Deposit date: | 1997-11-26 | | Release date: | 1999-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of bikunin from the inter-alpha-inhibitor complex: a serine protease inhibitor with two Kunitz domains.
J.Mol.Biol., 276, 1998
|
|
5F9Q
 
 | | Crystal structure of the extracellular domain of noncanonic ABC-type transporter YknZ from Gram-positive bacteria | | Descriptor: | Macrolide export ATP-binding/permease protein YknZ | | Authors: | Xu, Y, Guo, J, Jiang, R, Jin, X, Fan, S, Quan, C.S, Ha, N.C. | | Deposit date: | 2015-12-10 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | The Crystal Structure of the YknZ Extracellular Domain of ABC Transporter YknWXYZ from Bacillus amyloliquefaciens.
Plos One, 11, 2016
|
|
3K7D
 
 | | C-terminal (adenylylation) domain of E.coli Glutamine Synthetase Adenylyltransferase | | Descriptor: | Glutamate-ammonia-ligase adenylyltransferase, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2009-10-12 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Adenylylation Domain of E. coli Glutamine Synthetase Adenylyl Transferase: Evidence for Gene Duplication and Evolution of a New Active Site.
J.Mol.Biol., 396, 2010
|
|
8J86
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*TP*)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-04-30 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8G
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and cidofovir diphosphate | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
