5WHR
 
 | | Discovery of a novel and selective IDO-1 inhibitor PF-06840003 and its characterization as a potential clinical candidate. | | Descriptor: | (3R)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Greasley, S.E, Kaiser, S.E, Feng, J.L, Stewart, A. | | Deposit date: | 2017-07-18 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a Novel and Selective Indoleamine 2,3-Dioxygenase (IDO-1) Inhibitor 3-(5-Fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione (EOS200271/PF-06840003) and Its Characterization as a Potential Clinical Candidate.
J. Med. Chem., 60, 2017
|
|
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
3PRF
 
 | | Crystal Structure of Human B-Raf Kinase Domain in Complex with a Non-Oxime Furopyridine Inhibitor | | Descriptor: | 2-chloro-5-{[2-(pyrimidin-2-yl)furo[2,3-c]pyridin-3-yl]amino}phenol, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Vigers, G.P.A, Morales, T, Brandhuber, B.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Non-oxime inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PRI
 
 | | Crystal Structure of Human B-Raf Kinase in Complex with a Non-Oxime Furopyridine Inhibitor | | Descriptor: | 3-(4-{[2-(pyrimidin-2-yl)furo[2,3-c]pyridin-3-yl]amino}-1H-indazol-3-yl)propan-1-ol, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Vigers, G.P.A, Morales, T, Brandhuber, B.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Non-oxime inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PPJ
 
 | | Human B-Raf Kinase in Complex with a Furopyridine Inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, methyl 3-{[(5S)-1-(hydroxyamino)-5H-inden-5-yl]amino}furo[2,3-c]pyridine-2-carboxylate | | Authors: | Voegtli, W.C, Vigers, G.P.A, Morales, T, Brandhuber, B.J. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Non-oxime inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PPK
 
 | | Human B-Raf Kinase in Complex with a Non-Oxime Furopyridine Inhibitor | | Descriptor: | 3-[(5-hydroxynaphthalen-2-yl)amino]-N-(pyrimidin-4-yl)furo[2,3-c]pyridine-2-carboxamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Vigers, G.P.A, Morales, T, Brandhuber, B.J. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Non-oxime inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PSB
 
 | | Furo[2,3-c]pyridine-based Indanone Oximes as Potent and Selective B-Raf Inhibitors | | Descriptor: | B-RAF PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE, ethyl 3-{[1-(hydroxyamino)-2H-inden-5-yl]amino}thieno[2,3-c]pyridine-2-carboxylate | | Authors: | Morales, T, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-12-01 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Discovery of furo[2,3-c]pyridine-based indanone oximes as potent and selective B-Raf inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RR4
 
 | | tRNA-Guanine Transglycosylase in complex with N-Methyl-lin-Benzoguanine Inhibitor | | Descriptor: | 2,6-bis(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-04-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
3S1G
 
 | | tRNA-Guanine Transglycosylase in complex with lin-Benzohypoxanthine Inhibitor | | Descriptor: | 2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
6IIC
 
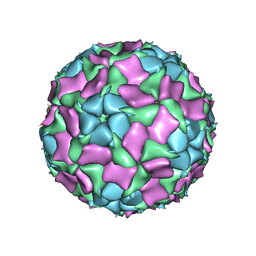 | | CryoEM structure of Mud Crab Dicistrovirus | | Descriptor: | VP1 of Mud crab dicistrovirus, VP2 of Mud crab dicistrovirus, VP3 of Mud crab dicistrovirus, ... | | Authors: | Zhang, Q, Gao, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Novel Viruses from Mud CrabScylla paramamosainwith Multiple Infections.
J. Virol., 93, 2019
|
|
7XR2
 
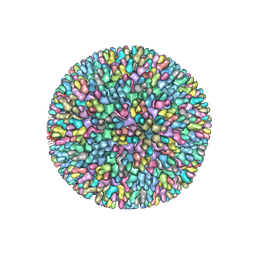 | |
7XR3
 
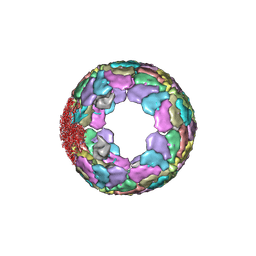 | |
6IZL
 
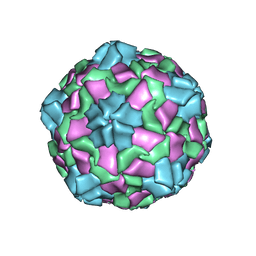 | |
4E4X
 
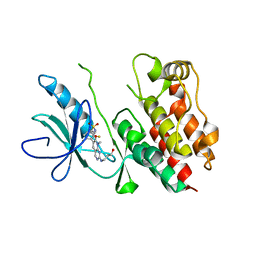 | | Crystal Structure of B-Raf Kinase Domain in Complex with a Dihydropyrido[2,3-d]pyrimidinone-based Inhibitor | | Descriptor: | N-(2,4-difluoro-3-{2-[(3-hydroxypropyl)amino]-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl}phenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C, Sturgis, H.L. | | Deposit date: | 2012-03-13 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The discovery of potent and selective pyridopyrimidin-7-one based inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2WVR
 
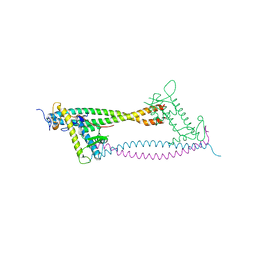 | | Human Cdt1:Geminin complex | | Descriptor: | DNA REPLICATION FACTOR CDT1, GEMININ | | Authors: | De Marco, V, Perrakis, A. | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Quaternary Structure of the Human Cdt1-Geminin Complex Regulates DNA Replication Licensing.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7U07
 
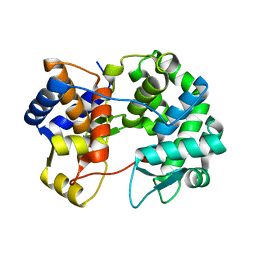 | |
7U1O
 
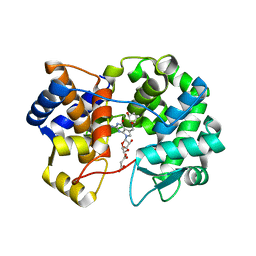 | | Crystal structure of queuine salvage enzyme DUF2419 complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U5A
 
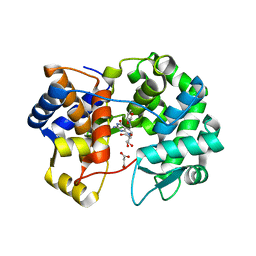 | | Crystal structure of queuine salvage enzyme DUF2419 mutant K199C, complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, MALONATE ION, Queuine salvage enzyme DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
3SM0
 
 | | tRNA-Guanine Transglycosylase in complex with lin-Benzohypoxanthine Inhibitor | | Descriptor: | 4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-06-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
7UK3
 
 | |
7U91
 
 | | Crystal structure of queuine salvage enzyme DUF2419, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7ULC
 
 | | Crystal structure of queuine salvage enzyme DUF2419 mutant D231N, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, Queuosine salvage protein DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-04-04 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7UGK
 
 | |
3TLL
 
 | | tRNA-Guanine Transglycosylase in complex with N-Ethyl-lin-benzoguanine Inhibitor | | Descriptor: | 6-(ethylamino)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Klebe, G, Immekus, F, Heine, A. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | From lin-Benzoguanines to lin-Benzohypoxanthines as Ligands for Zymomonas mobilis tRNA-Guanine Transglycosylase: Replacement of Protein-Ligand Hydrogen Bonding by Importing Water Clusters.
Chemistry, 18, 2012
|
|
