2V6E
 
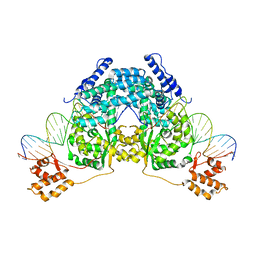 | |
1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
2V0O
 
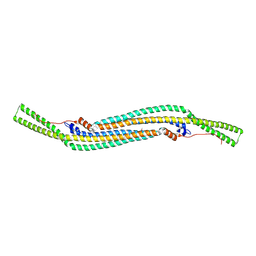 | | FCHO2 F-BAR domain | | Descriptor: | ACETATE ION, FCH DOMAIN ONLY PROTEIN 2 | | Authors: | Henne, W.M, McMahon, H.T, Kent, H.M, Evans, P.R. | | Deposit date: | 2007-05-15 | | Release date: | 2007-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Analysis of Fcho2 F-Bar Domain: A Dimerizing and Membrane Recruitment Module that Effects Membrane Curvature.
Structure, 15, 2007
|
|
1BRV
 
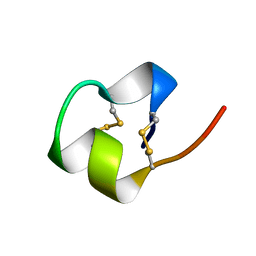 | | SOLUTION NMR STRUCTURE OF THE IMMUNODOMINANT REGION OF PROTEIN G OF BOVINE RESPIRATORY SYNCYTIAL VIRUS, 48 STRUCTURES | | Descriptor: | PROTEIN G | | Authors: | Doreleijers, J.F, Langedijk, J.P.M, Hard, K, Rullmann, J.A.C, Boelens, R, Schaaper, W.M, Van Oirschot, J.T, Kaptein, R. | | Deposit date: | 1996-03-29 | | Release date: | 1997-06-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant region of protein G of bovine respiratory syncytial virus.
Biochemistry, 35, 1996
|
|
2V4B
 
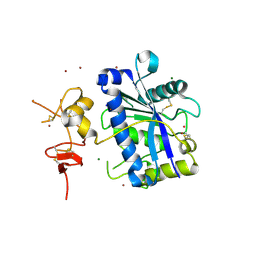 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
1QD7
 
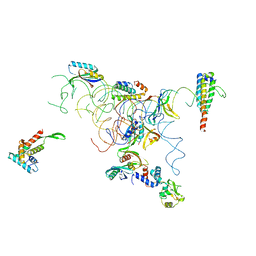 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
4B8D
 
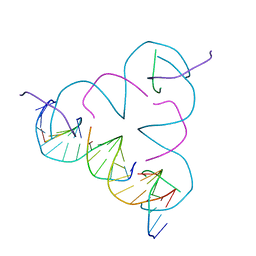 | | TENSEGRITY TRIANGLE FROM ENZYMATICALLY MANUFACTURED DNA | | Descriptor: | 5'-D(*CP*CP*GP*TP*AP*CP*AP)-3', 5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*DGP *GP*AP*CP*AP*TP*CP*A)-3', 5'-D(*GP*GP*CP*TP*GP*CP)-3', ... | | Authors: | Ducani, C, Kaul, C.D, Moche, M, Shih, W.M, Hogberg, B. | | Deposit date: | 2012-08-26 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.79 Å) | | Cite: | Enzymatic Production of 'Monoclonal Stoichiometric' Single-Stranded DNA Oligonucleotides
Nat.Methods, 10, 2013
|
|
2XCN
 
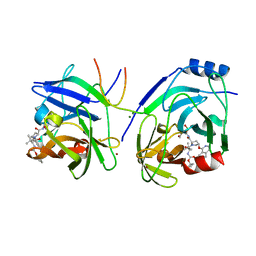 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | MAGNESIUM ION, N-[(CYCLOPENTYLOXY)CARBONYL]-3-METHYL-L-VALYL-(4R)-N-{(1R)-3-HYDROXY-1-[HYDROXY(OXIDO)BORANYL]PROPYL}-4-(ISOQUINOLIN-1-YLOXY)-L-PROLINAMIDE, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XCF
 
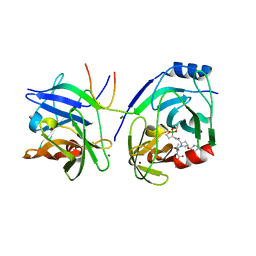 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | CYCLOPENTYL N-[(2S)-1-[(2S,4R)-2-[[(4R)-8-HYDROXY-1,6,10-TRIOXA-5$L^{4}-BORASPIRO[4.5]DECAN-4-YL]CARBAMOYL]-4-ISOQUINOLIN-1-YLOXY-PYRROLIDIN-1-YL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2VLE
 
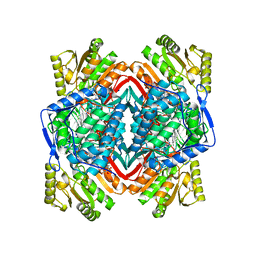 | | The structure of daidzin, a naturally occurring anti alcohol- addiction agent, in complex with human mitochondrial aldehyde dehydrogenase | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL, DAIDZIN | | Authors: | Lowe, E.D, Gao, G.Y, Johnson, L.N, Keung, W.M. | | Deposit date: | 2008-01-13 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Daidzin, a Naturally Occurring Anti-Alcohol-Addiction Agent, in Complex with Human Mitochondrial Aldehyde Dehydrogenase.
J.Med.Chem., 51, 2008
|
|
3ZBH
 
 | |
4AQW
 
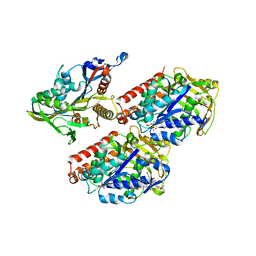 | | Model of human kinesin-5 motor domain (1II6, 3HQD) and mammalian tubulin heterodimer (1JFF) docked into the 9.5-angstrom cryo-EM map of microtubule-bound kinesin-5 motor domain in the rigor state. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Goulet, A, Behnke-Parks, W.M, Sindelar, C.V, Rosenfeld, S.S, Moores, C.A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Structural Basis of Force Generation by the Mitotic Motor Kinesin-5.
J.Biol.Chem., 287, 2012
|
|
4AQV
 
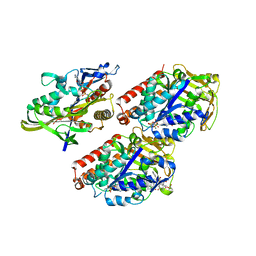 | | Model of human kinesin-5 motor domain (3HQD) and mammalian tubulin heterodimer (1JFF) docked into the 9.7-angstrom cryo-EM map of microtubule-bound kinesin-5 motor domain in the AMPPPNP state. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Goulet, A, Behnke-Parks, W.M, Sindelar, C.V, Rosenfeld, S.S, Moores, C.A. | | Deposit date: | 2012-04-19 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | The Structural Basis of Force Generation by the Mitotic Motor Kinesin-5.
J.Biol.Chem., 287, 2012
|
|
2VXS
 
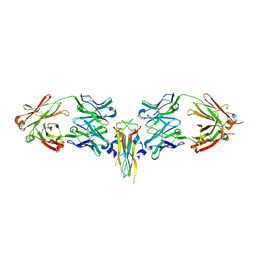 | | Structure of IL-17A in complex with a potent, fully human neutralising antibody | | Descriptor: | FAB FRAGMENT, INTERLEUKIN-17A, SULFATE ION | | Authors: | Gerhardt, S, Hargreaves, D, Pauptit, R.A, Davies, R.A, Russell, C, Welsh, F, Tuske, S.J, Coales, S.J, Hamuro, Y, Needham, M.R.C, Langham, C, Barker, W, Bell, P, Aziz, A, Smith, M.J, Dawson, S, Abbott, W.M. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of Il-17A in Complex with a Potent, Fully Human Neutralising Antibody.
J.Mol.Biol., 394, 2009
|
|
1DAV
 
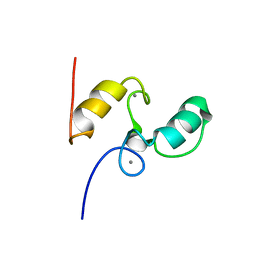 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (20 STRUCTURES) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
1DAQ
 
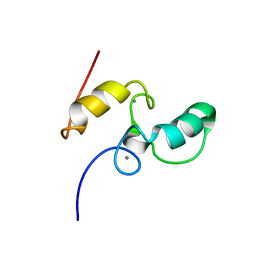 | | SOLUTION STRUCTURE OF THE TYPE I DOCKERIN DOMAIN FROM THE CLOSTRIDIUM THERMOCELLUM CELLULOSOME (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | CALCIUM ION, ENDOGLUCANASE SS | | Authors: | Lytle, B.L, Volkman, B.F, Westler, W.M, Heckman, M.P, Wu, J.H.D. | | Deposit date: | 1999-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain.
J.Mol.Biol., 307, 2001
|
|
1SJG
 
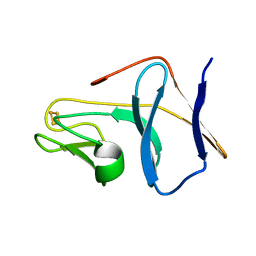 | | Solution Structure of T4moC, the Rieske Ferredoxin Component of the Toluene 4-Monooxygenase Complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Toluene-4-monooxygenase system protein C | | Authors: | Skjeldal, L, Peterson, F.C, Doreleijers, J.F, Moe, L.A, Pikus, J.D, Volkman, B.F, Westler, W.M, Markley, J.L, Fox, B.G. | | Deposit date: | 2004-03-03 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of T4moC, the Rieske ferredoxin component of the toluene 4-monooxygenase complex
J.Biol.Inorg.Chem., 9, 2004
|
|
6Q1W
 
 | |
6OV9
 
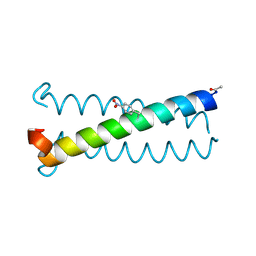 | |
6OVS
 
 | | Coiled-coil Trimer with Glu:Tyr:Lys Triad | | Descriptor: | Coiled-coil Trimer with Glu:Tyr:Lys Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-05-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
4WWR
 
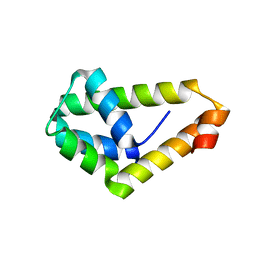 | |
6OVV
 
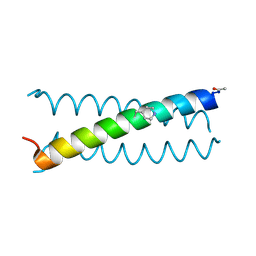 | |
6Q22
 
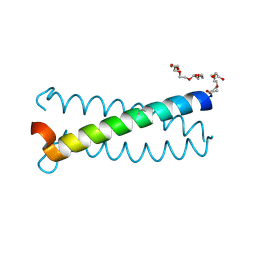 | | Coiled-coil Trimer with Glu:Aminobutyric acid:Lys Triad | | Descriptor: | Coiled-coil Trimer with Glu:Aminobutyric acid:Lys Triad, PENTAETHYLENE GLYCOL | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-08-06 | | Release date: | 2020-04-29 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
6Q25
 
 | |
7WD4
 
 | | Crystal structure of the Ilheus virus helicase: implications for enzyme function and drug design | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NS3 helicase | | Authors: | Wang, D.P, Wang, M.Y, Zhou, X, Wang, W.M, Cao, J.M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the Ilheus virus helicase: implications for enzyme function and drug design
To Be Published
|
|
