4WXB
 
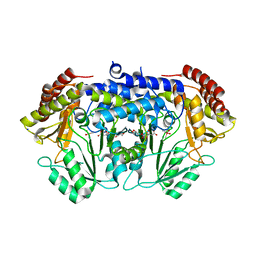 | | Crystal Structure of Serine Hydroxymethyltransferase from Streptococcus thermophilus | | Descriptor: | CACODYLATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wandtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | Deposit date: | 2014-11-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4YTB
 
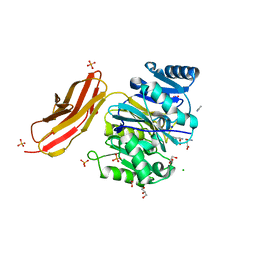 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in complex with dipeptide Asp-Gln. | | Descriptor: | ASPARTIC ACID, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
4YTG
 
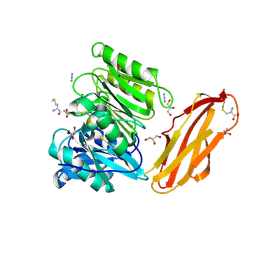 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) mutant C351A in complex with dipeptide Met-Arg. | | Descriptor: | ARGININE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
3DKX
 
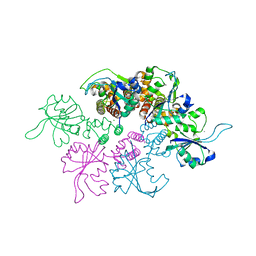 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), trigonal form, to 2.7 Ang resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
3DKY
 
 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), tetragonal form, to 3.6 Ang resolution | | Descriptor: | MANGANESE (II) ION, Replication protein repB | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
1S9B
 
 | | Crystal Structure Analysis of the B-DNA GAATTCG | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*CP*G)-3', NICKEL (II) ION | | Authors: | Valls, N, Uson, I, Gouyette, C, Subirana, J.A. | | Deposit date: | 2004-02-04 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A cubic arrangement of DNA double helices based on nickel-guanine interactions
J.Am.Chem.Soc., 126, 2004
|
|
1VZ5
 
 | | Succinate Complex of AtsK | | Descriptor: | PUTATIVE ALKYLSULFATASE ATSK, SUCCINIC ACID | | Authors: | Mueller, I, Stueckl, A.C, Uson, I, Kertesz, M. | | Deposit date: | 2004-05-14 | | Release date: | 2004-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Succinate Complex Crystal Structures of the Alpha-Ketoglutarate-Dependent Dioxygenase Atsk: Steric Aspects of Enzyme Self-Hydroxylation
J.Biol.Chem., 280, 2005
|
|
6X8O
 
 | | BimBH3 peptide tetramer | | Descriptor: | Bcl-2-like protein 11, THIOCYANATE ION | | Authors: | Robin, A.Y, Westphal, D, Uson, I, Czabotar, P.E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biophysical Characterization of Pro-apoptotic BimBH3 Peptides Reveals an Unexpected Capacity for Self-Association.
Structure, 29, 2021
|
|
1VZ4
 
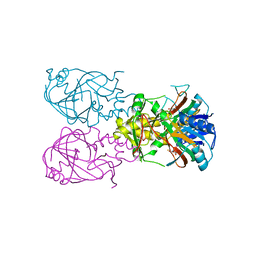 | | Fe-Succinate Complex of AtsK | | Descriptor: | FE (II) ION, PUTATIVE ALKYLSULFATASE ATSK, SUCCINIC ACID | | Authors: | Mueller, I, Stueckl, A.C, Uson, I, Kertesz, M. | | Deposit date: | 2004-05-14 | | Release date: | 2004-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Succinate Complex Crystal Structures of the Alpha-Ketoglutarate-Dependent Dioxygenase Atsk: Steric Aspects of Enzyme Self-Hydroxylation
J.Biol.Chem., 280, 2005
|
|
6YRW
 
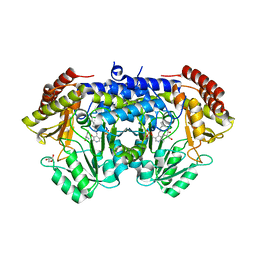 | | SHMT from Streptococcus thermophilus Tyr55Ser variant as internal aldimine and as non-covalent complex with D-Ser | | Descriptor: | D-SERINE, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2020-04-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into nucleophile substrate specificity in variants of N-Serine hydroxymethyltransferase from Streptococcus thermophilus
To Be Published
|
|
4LUN
 
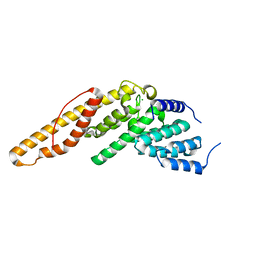 | | Structure of the N-terminal mIF4G domain from S. cerevisiae Upf2, a protein involved in the degradation of mRNAs containing premature stop codons | | Descriptor: | CHLORIDE ION, Nonsense-mediated mRNA decay protein 2 | | Authors: | Fourati, Z, Roy, B, Millan, C, Courreux, P.D, Kervestin, S, van Tilbeurgh, H, He, F, Uson, I, Jacobson, A, Graille, M. | | Deposit date: | 2013-07-25 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | A highly conserved region essential for NMD in the Upf2 N-terminal domain.
J.Mol.Biol., 426, 2014
|
|
4MIV
 
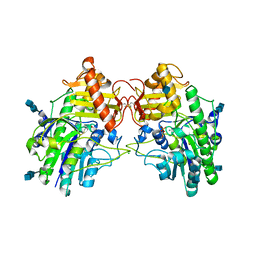 | | Crystal Structure of Sulfamidase, Crystal Form L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Sheldrick, G.M, Gaertner, J, Kraetzner, R, Steinfeld, R. | | Deposit date: | 2013-09-02 | | Release date: | 2014-05-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5NDX
 
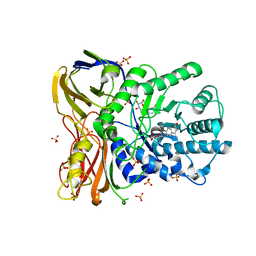 | | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent: Crystallography and QM/MM insights into Mucopolysaccharidosis I | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyl hydrolase, SULFATE ION | | Authors: | Raich, L, Valero-Gonzalez, J, Castro-Lopez, J, Millan, C, Jimenez-Garcia, M.J, Nieto, P, Uson, I, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2017-03-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent:
Crystallography and QM/MM insights into Mucopolysaccharidosis I.
To Be Published
|
|
4MHX
 
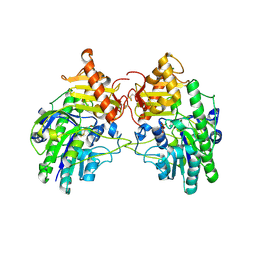 | | Crystal Structure of Sulfamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Gaertner, J, Kraetzner, R, Steinfeld, R, Sheldrick, G.M. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3L0O
 
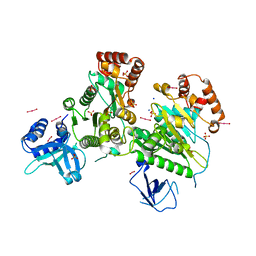 | | Structure of RNA-free Rho transcription termination factor from Thermotoga maritima | | Descriptor: | SODIUM ION, SULFATE ION, Transcription termination factor rho, ... | | Authors: | Canals, A, Uson, I, Coll, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of RNA-Free Rho Termination Factor Indicates a Dynamic Mechanism of Transcript Capture
J.Mol.Biol., 400, 2010
|
|
2ET0
 
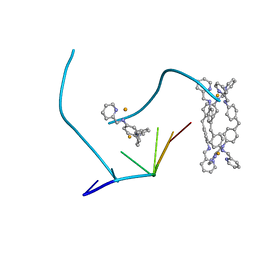 | | The structure of a three-way DNA junction in complex with a metallo-supramolecular helicate reveals a new target for drugs | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Oleksi, A, Blanco, A.G, Boer, R, Uson, I, Aymami, J, Coll, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Recognition of a Three-Way DNA Junction by a Metallosupramolecular Helicate
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
3N6S
 
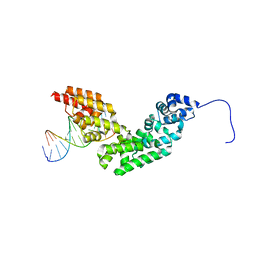 | | Crystal structure of human mitochondrial mTERF in complex with a 15-mer DNA encompassing the tRNALeu(UUR) binding sequence | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*A)-3'), Transcription termination factor, ... | | Authors: | Jimenez-Menendez, N, Fernandez-Millan, P, Rubio-Cosials, A, Arnan, C, Montoya, J, Jacobs, H.T, Bernado, P, Coll, M, Uson, I, Sola, M. | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3N6T
 
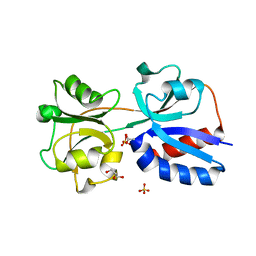 | | Effector binding domain of TsaR | | Descriptor: | ACETATE ION, GLYCEROL, LysR type regulator of tsaMBCD, ... | | Authors: | Monferrer, D, Uson, I. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies on the effector binding domain of TsaR
To be Published
|
|
3N7Q
 
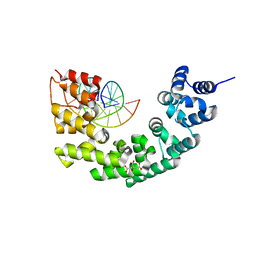 | | Crystal structure of human mitochondrial mTERF fragment (aa 99-399) in complex with a 12-mer DNA encompassing the tRNALeu(UUR) binding sequence | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*G)-3'), ... | | Authors: | Jimenez-Menendez, N, Fernandez-Millan, P, Rubio-Cosials, A, Arnan, C, Montoya, J, Jacobs, H.T, Bernado, P, Coll, M, Uson, I, Sola, M. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3N6U
 
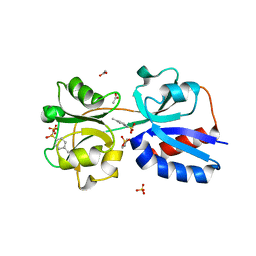 | |
1HDH
 
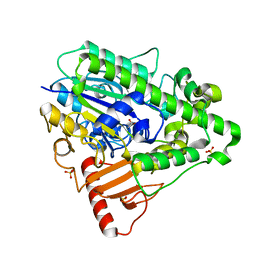 | | Arylsulfatase from Pseudomonas aeruginosa | | Descriptor: | Arylsulfatase, CALCIUM ION, SULFATE ION | | Authors: | Boltes, I, Czapinska, H, Kahnert, A, von Buelow, R, Dirks, T, Schmidt, B, von Figura, K, Kertesz, M.A, Uson, I. | | Deposit date: | 2000-11-16 | | Release date: | 2001-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 A Structure of Arylsulfatase from Pseudomonas Aeruginosa Establishes the Catalytic Mechanism of Sulfate Ester Cleavage in the Sulfatase Family.
Structure, 9, 2001
|
|
1OVN
 
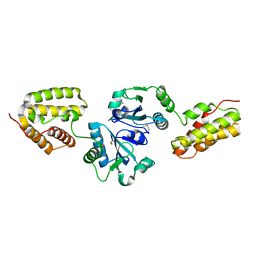 | | Crystal Structure and Functional Analysis of Drosophila Wind-- a PDI-Related Protein | | Descriptor: | CESIUM ION, Windbeutel | | Authors: | Ma, Q, Guo, C, Barnewitz, K, Sheldrick, G.M, Soling, H.D, Uson, I, Ferrari, D.M. | | Deposit date: | 2003-03-27 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of Drosophila Wind, a protein-disulfide isomerase-related protein.
J.Biol.Chem., 278, 2003
|
|
3GNG
 
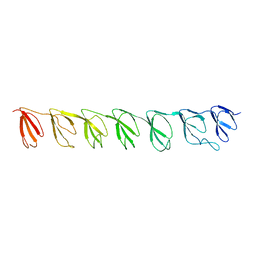 | | P21B crystal structure of R1-R7 of Murine MVP | | Descriptor: | Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Uson, I, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
3HCL
 
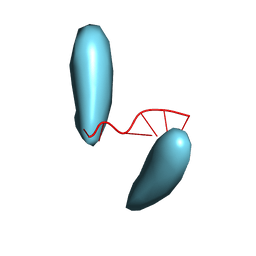 | | Helical superstructures in a DNA oligonucleotide crystal | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*AP*T)-3') | | Authors: | De Luchi, D, Martinez de Ilarduya, I, Subirana, J.A, Uson, I, Campos, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A geometric approach to the crystallographic solution of nonconventional DNA structures: helical superstructures of d(CGATAT)
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3GNF
 
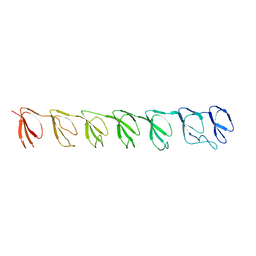 | | P1 Crystal structure of the N-terminal R1-R7 of murine MVP | | Descriptor: | Major vault protein | | Authors: | Querol-Audi, J, Casanas, A, Uson, I, Luque, D, Caston, J.R, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP
Embo J., 28, 2009
|
|
