2XAX
 
 | |
2X0X
 
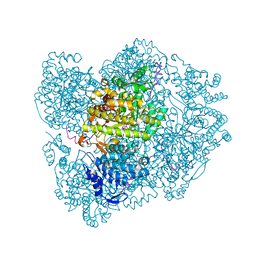 | | Ribonucleotide reductase R1 subunit of E. coli to 2.3 A resolution | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA, SULFATE ION | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-Specific Incorporation of 3-Nitrotyrosine as a Probe of Pk(A) Perturbation of Redox-Active Tyrosines in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
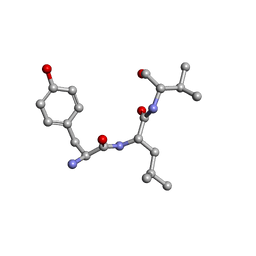
1GJ1
 
 | | NMR structure of d(CCAAAGXACTGGG), X is a 3'phosphoglycolate, 5'phosphate gapped lesion | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 5'-D(*CP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*CP*AP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3', ... | | Authors: | Junker, H.-D, Hoehn, S.T, Bunt, R.C, Marathius, V, Chen, J, Turner, C.J, Stubbe, J. | | Deposit date: | 2002-10-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Synthesis, Characterization and Solution Structure of Tethered Oligonucleotides Containing an Internal 3'-Phosphoglycolate, 5'-Phosphate Gapped Lesion
Nucleic Acids Res., 30, 2002
|
|
1TWJ
 
 | |
2XAK
 
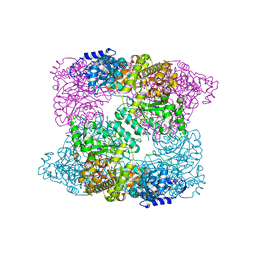 | | Ribonucleotide reductase Y730NO2Y modified R1 subunit of E. coli | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Site-Specific Incorporation of 3-Nitrotyrosine as a Probe of Pk(A) Perturbation of Redox-Active Tyrosines in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
2XAP
 
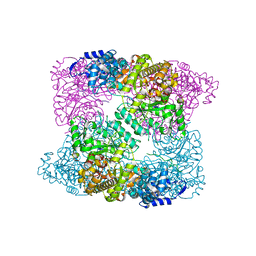 | |
1G5E
 
 | |
1G5D
 
 | |
1GJ0
 
 | |
1GIZ
 
 | |
1GSO
 
 | |
1QCZ
 
 | |
2AV8
 
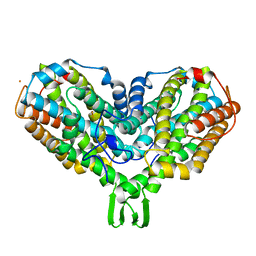 | | Y122F MUTANT OF RIBONUCLEOTIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | FE (II) ION, MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
4U3E
 
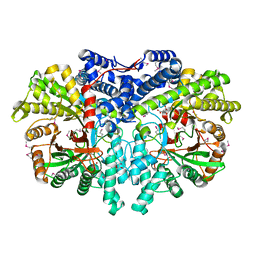 | | Anaerobic ribonucleotide reductase | | Descriptor: | ACETATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-07-20 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The class III ribonucleotide reductase from Neisseria bacilliformis can utilize thioredoxin as a reductant.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9DB2
 
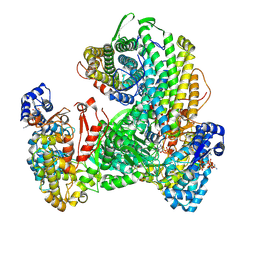 | | Class Ia ribonucleotide reductase with mechanism-based inhibitor N3CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 4-amino-1-{(3xi)-3-C-amino-2-deoxy-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-beta-D-threo-pentofuranosyl}pyrimidin-2(1H)-one, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Westmoreland, D.E, Drennan, C.L. | | Deposit date: | 2024-08-23 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | 2.6- angstrom resolution cryo-EM structure of a class Ia ribonucleotide reductase trapped with mechanism-based inhibitor N 3 CDP.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3HJZ
 
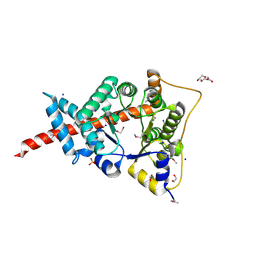 | | The structure of an aldolase from Prochlorococcus marinus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5T6O
 
 | |
3D54
 
 | | Structure of PurLQS from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Formylglycinamide ribonucleotide amidotransferase, Phosphoribosylformylglycinamidine synthase 1, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2008-05-15 | | Release date: | 2008-07-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima: structural insights into complex formation.
Biochemistry, 47, 2008
|
|
6AUI
 
 | | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Brignole, E.J, Drennan, C.L, Asturias, F.J, Tsai, K.L, Penczek, P.A. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3- angstrom resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound.
Elife, 7, 2018
|
|
4N82
 
 | | X-ray crystal structure of Streptococcus sanguinis NrdIox | | Descriptor: | FLAVIN MONONUCLEOTIDE, Ribonucleotide reductase, SULFATE ION | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2013-10-16 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Streptococcus sanguinis Class Ib Ribonucleotide Reductase: HIGH ACTIVITY WITH BOTH IRON AND MANGANESE COFACTORS AND STRUCTURAL INSIGHTS.
J.Biol.Chem., 289, 2014
|
|
4N83
 
 | |
3UUS
 
 | |
6CGM
 
 | |
6CGL
 
 | |
6CGN
 
 | |
