1GI1
 
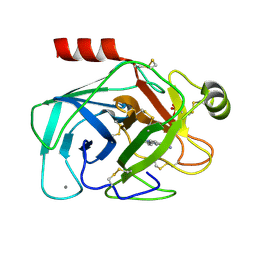 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI0
 
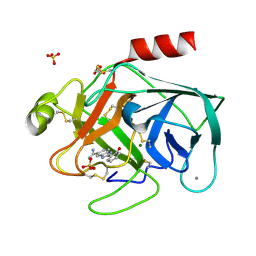 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GHZ
 
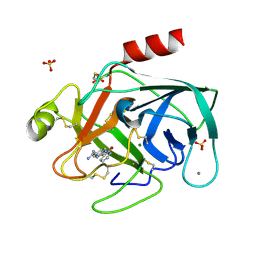 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI5
 
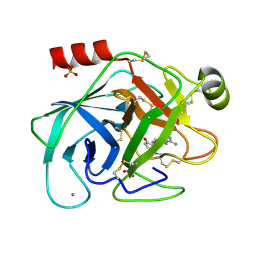 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-5-METHOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GHX
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI7
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GHY
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(3-HYDROXY-PYRIDIN-2-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI4
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-3H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GHW
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site
J.Mol.Biol., 307, 2001
|
|
1GI8
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI3
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
7AFZ
 
 | | L1 metallo-b-lactamase with compound EBL-1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors.
Nat.Chem., 14, 2022
|
|
8BW3
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2S)-N-(cyclopropylmethyl)-2-methyl-4-(1-methyl-1H-pyrrole-2-carbonyl)piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW4
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-4-(3-fluoranylthiophen-2-yl)carbonyl-N-(4-methoxyphenyl)-2-methyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
8BW2
 
 | | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative | | Descriptor: | (2R)-N-(2-methoxyethyl)-2-methyl-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Hassell-Hart, S, Bradshaw, W.J, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Biggin, P.C, Spencer, J, von Delft, F. | | Deposit date: | 2022-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis -- Crystal Structure of PHIP in complex with Z198194396 synthetic derivative
To Be Published
|
|
5N5H
 
 | | Crystal structure of metallo-beta-lactamase VIM-1 in complex with ML302F inhibitor | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
5N5I
 
 | | Crystal Structure of VIM-1 metallo-beta-lactamase in complex with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
5N5G
 
 | |
1KQ7
 
 | | E315Q Mutant Form of Fumarase C from E.coli | | Descriptor: | CITRIC ACID, D-MALATE, FUMARATE HYDRATASE CLASS II | | Authors: | Weaver, T.M, Estevez, M, Skarda, J, Spencer, J. | | Deposit date: | 2002-01-04 | | Release date: | 2002-08-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystallographic and kinetic correlation of a clinically observed human fumarase mutation.
Protein Sci., 11, 2002
|
|
1SML
 
 | | METALLO BETA LACTAMASE L1 FROM STENOTROPHOMONAS MALTOPHILIA | | Descriptor: | PROTEIN (PENICILLINASE), ZINC ION | | Authors: | Ullah, J.H, Walsh, T.R, Taylor, I.A, Emery, D.C, Verma, C.S, Gamblin, S.J, Spencer, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia at 1.7 A resolution.
J.Mol.Biol., 284, 1998
|
|
3SD9
 
 | |
8R5T
 
 | |
8R5U
 
 | |
4U4L
 
 | | Crystal structure of the metallo-beta-lactamase NDM-1 in complex with a bisthiazolidine inhibitor | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase NDM-1, GLYCEROL, ... | | Authors: | Kosmopoulou, M, Hinchliffe, P, Spencer, J. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase NDM-1 in complex with a bisthiazolidine inhibitor
To Be Published
|
|
8R2Z
 
 | | Structure of CTX-M-15 complexed with benzoxaborole AK-425 | | Descriptor: | (9S)-7-oxidanyl-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-triene-9-carbonitrile, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2023-11-08 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of CTX-M-15 complexed with benzoxaborole AK-425
To Be Published
|
|
