5F4R
 
 | | HIV-1 gp120 complex with BNW-IV-147 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, FORMIC ACID, ... | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
ACS Med Chem Lett, 7, 2016
|
|
5F4P
 
 | | HIV-1 gp120 complex with BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-5-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
Acs Med.Chem.Lett., 7, 2016
|
|
5F4L
 
 | | HIV-1 gp120 complex with JP-III-048 | | Descriptor: | ENVELOPE GLYCOPROTEIN GP120 of HIV-1 clade C, ~{N}'-[(1~{R},2~{R})-2-(carbamimidamidomethyl)-6-(methylaminomethyl)-2,3-dihydro-1~{H}-inden-1-yl]-~{N}-(4-chloranyl-3-fluoranyl-phenyl)ethanediamide | | Authors: | Liang, S, Hendrickson, W.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-Molecule CD4-Mimics: Structure-Based Optimization of HIV-1 Entry Inhibition.
Acs Med.Chem.Lett., 7, 2016
|
|
4I54
 
 | |
4I53
 
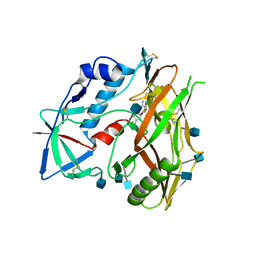 | |
7N6W
 
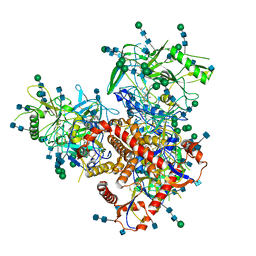 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U2 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
7N6U
 
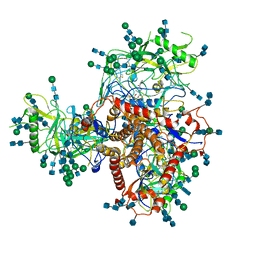 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U1 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, S, Wang, K, Mao, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-09-22 | | Last modified: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
6ONE
 
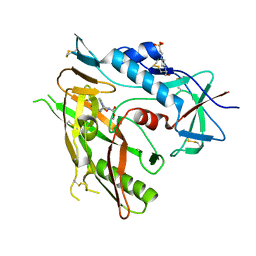 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-188-A01. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, clade A/E 93TH057 HIV-1 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6ONH
 
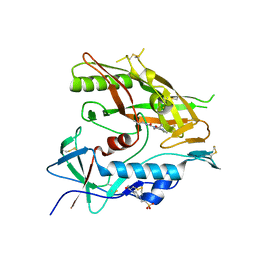 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-IV-031-A05. | | Descriptor: | (3S)-N~3~-(4-chloro-3-fluorophenyl)-N~1~-propylpiperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6ONV
 
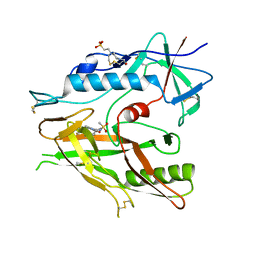 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-027-D05. | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-(methylsulfonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6MUG
 
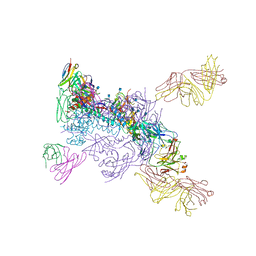 | |
6MU7
 
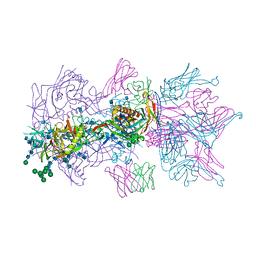 | |
6MTJ
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies 3H109L and 35O22 at 2.9 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-19 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6MUF
 
 | |
6MTN
 
 | |
6MU8
 
 | |
6MU6
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-814508 in Complex with Human Antibodies 3H109L and 35O22 at 3.2 Angstrom | | Descriptor: | (2R)-{1-[{7-[2-({[3-(dimethylamino)propyl](methyl)amino}methyl)-1,3-thiazol-4-yl]-4-methoxy-1H-pyrrolo[2,3-c]pyridin-3-yl}(oxo)acetyl]piperidin-4-yl}(phenyl)acetonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
