5TGD
 
 | |
1TYP
 
 | |
1TYT
 
 | |
5HHG
 
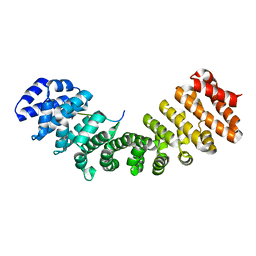 | | Mouse importin alpha: Dengue 2 NS5 C-terminal NLS peptide complex | | Descriptor: | Importin subunit alpha-1, RNA-directed RNA polymerase NS5 | | Authors: | Smith, K.M, Forwood, J.K. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production.
PLoS Pathog., 12, 2016
|
|
1P5W
 
 | |
1P5Y
 
 | |
2PR9
 
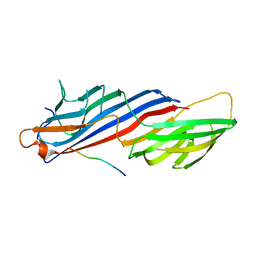 | | Mu2 adaptin subunit (AP50) of AP2 adaptor (second domain), complexed with GABAA receptor-gamma2 subunit-derived internalization peptide DEEYGYECL | | Descriptor: | AP-2 complex subunit mu-1, GABA(A) receptor subunit gamma-2 peptide | | Authors: | Vahedi-Faridi, A, Haucke, V, Kittler, J.T, Kukhtina, V, Moss, S.J, Saenger, W, Chen, G.-J, Tretter, V, Smith, K, Yan, Z, McAinsh, K, Arancibia-Carcamo, L. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5BVK
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | 1-(2-chlorophenyl)-3-(pyridin-3-ylmethyl)urea, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5BVO
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(5-{(1S)-1-[(5-fluoro-1,3-benzoxazol-2-yl)amino]ethyl}-2-methylphenyl)imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5BVW
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5BVN
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-[5-({[(3-fluorophenyl)carbamoyl]amino}methyl)-2-methylphenyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4HHA
 
 | | Anti-Human Cytomegalovirus (HCMV) Fab KE5 with epitope peptide AD-2S1 | | Descriptor: | Antibody KE5, CHLORIDE ION, Fab KE5, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Pfoh, R, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HH9
 
 | | Anti-Human Cytomegalovirus (HCMV) Fab KE5 | | Descriptor: | Fab KE5, heavy chain, light chain | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Pfoh, R, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HIE
 
 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 | | Descriptor: | Antibody 023.102, Fab 023.102 | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HIJ
 
 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound L-rhamnose-(1-2)-alpha-D-galactose-(3-O)-phosphate-2-glycerol | | Descriptor: | Fab 023.102 heavy chain, Fab 023.102 light chain, GLYCEROL, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HII
 
 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound rhamnose-galactose | | Descriptor: | Fab 023.102 heavy chain, Fab 023.102 light chain, alpha-L-rhamnopyranose-(1-2)-beta-D-galactopyranose | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
4HIH
 
 | | Anti-Streptococcus pneumoniae 23F Fab 023.102 with bound rhamnose. | | Descriptor: | Antibody 023.102, Fab 023.102, alpha-L-rhamnopyranose | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
3G83
 
 | |
3G84
 
 | |
3G81
 
 | |
7JMU
 
 | | Hen egg-white lysozyme with ionic liquid ethylammonium nitrate | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Greaves, T, Drummond, C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Lysozyme conformational changes with ionic liquids: Spectroscopic, small angle x-ray scattering and crystallographic study.
J Colloid Interface Sci, 585, 2021
|
|
1FPV
 
 | |
2CAS
 
 | |
3DBZ
 
 | | human surfactant protein D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Head, J.F. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of recombinant surfactant protein D with lipopolysaccharide: conformation and orientation of bound protein by IRRAS and simulations.
Biochemistry, 47, 2008
|
|
4DPV
 
 | | PARVOVIRUS/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*CP*TP*CP*TP*TP*GP*C)-3'), MAGNESIUM ION, PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Chapman, M.S, Rossmann, M.G. | | Deposit date: | 1996-02-01 | | Release date: | 1997-04-01 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Canine parvovirus capsid structure, analyzed at 2.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
