5Z4W
 
 | | Crystal structure of signalling protein from buffalo (SPB-40) with an altered conformation of Trp78 at 1.79 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Singh, P.K, Chaudhary, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-01-15 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A glycoprotein from mammary gland secreted during involution promotes apoptosis: Structural and biological studies.
Arch. Biochem. Biophys., 644, 2018
|
|
5XGY
 
 | | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Shokeen, A, Sharma, P, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-04-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution
To Be Published
|
|
5X47
 
 | |
6KL8
 
 | | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site
To Be Published
|
|
8HE6
 
 | | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution | | Descriptor: | All3014 protein, CALCIUM ION, MAGNESIUM ION | | Authors: | Chatterjee, A, Singh, P.K, Singh, T.P, Marina, A, Sharma, S, Rai, L.C. | | Deposit date: | 2022-11-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a fosfomycin and bleomycin resistant protein (ALL3014) from Anabaena/Nostoc cyanobacterium at 1.70 A resolution
To Be Published
|
|
6IDM
 
 | | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution
To Be Published
|
|
6J93
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase form apo at 0.95 A resolution.
To Be Published
|
|
5CRY
 
 | | Structure of iron-saturated C-lobe of bovine lactoferrin at pH 6.8 indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron-saturated C-lobe of bovine lactoferrin at pH 7.0 indicates the softening of iron coordination
To Be Published
|
|
6IF7
 
 | | Crystal Structure of AA10 Lytic Polysaccharide Monooxygenase from Tectaria macrodonta | | Descriptor: | COPPER (II) ION, Chitin binding protein, GLYCEROL, ... | | Authors: | Archana, A, Yadav, S.K, Singh, P.K, Vasudev, P.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase.
Planta, 249, 2019
|
|
5WRF
 
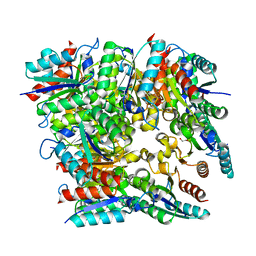 | | Crystal structure of dodecameric type II dehydroquinate dehydratase from Acinetobacter baumannii with unexplained connecting electron density between free cysteine residues of molecular pairs | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase | | Authors: | Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of dodecameric type II dehydroquinate dehydratase from Acinetobacter baumannii with unexplained connecting electron density between free cysteine residues of molecular pairs
To Be Published
|
|
2M6Y
 
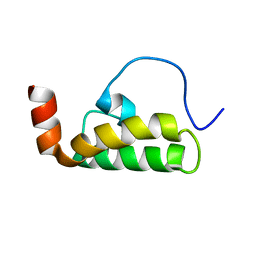 | | The solution structure of the J-domain of human DnaJA1 | | Descriptor: | DnaJ homolog subfamily A member 1 | | Authors: | Stark, J.L, Mehla, K, Chaika, N, Acton, T.B, Xiao, R, Singh, P.K, Montelione, G.T, Powers, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer.
Biochemistry, 53, 2014
|
|
5WV1
 
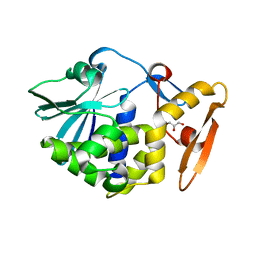 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at 1.90 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Shokeen, A, Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at
1.90 A resolution.
To Be Published
|
|
5TRS
 
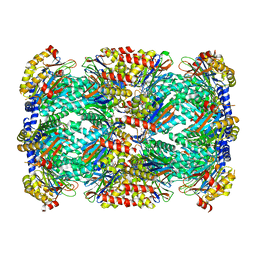 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRR
 
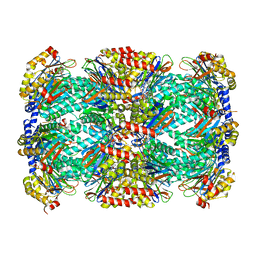 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRY
 
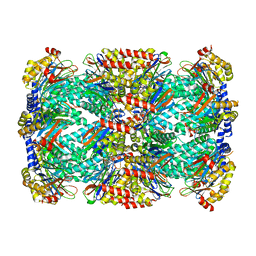 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TS0
 
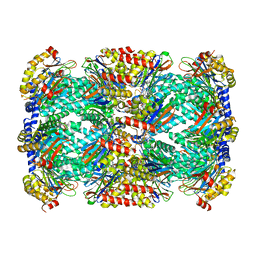 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2208 | | Descriptor: | (2S)-N-{(2S)-3-methoxy-1-[(naphthalen-1-ylmethyl)amino]-1-oxopropan-2-yl}-4-oxo-2-[(3-phenylpropanoyl)amino]-4-(1H-pyrrol-1-yl)butanamide (non-preferred name), Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84679747 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5HBC
 
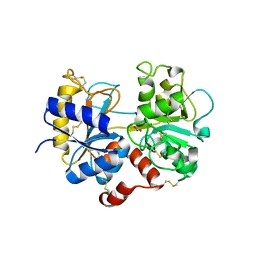 | | Intermediate structure of iron-saturated C-lobe of bovine lactoferrin at 2.79 Angstrom resolution indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-31 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron saturated C-lobe of bovine lactoferrin at pH 6.8 indicates a weakening of iron coordination
Proteins, 84, 2016
|
|
5GM7
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Ribosome inactivating protein | | Authors: | Singh, B, Singh, P.K, Pandey, S.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-07-13 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina at 1.78 Angstrom resolution
To Be Published
|
|
7WP3
 
 | | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, Proliferating cell nuclear antigen | | Authors: | Ahmad, M.I, Yadav, S.P, Singh, P.K, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-01-22 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution
To Be Published
|
|
7XFW
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.07 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the complex of camel peptidoglycan recognition protein-S with hexanoic acid reveals novel features of the versatile ligand-binding site at the dimeric interface.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
7XFX
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution.
To Be Published
|
|
7XFY
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution.
To Be Published
|
|
7XU8
 
 | | Structure of the complex of camel peptidoglycan recognition protein-short (PGRP-S) with heptanoic acid at 2.15 A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Maurya, A, Ahmad, N, Viswanathan, V, Singh, P.K, Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand recognition by peptidoglycan recognition protein-S (PGRP-S): structure of the complex of camel PGRP-S with heptanoic acid at 2.15 angstrom resolution.
Int J Biochem Mol Biol, 13, 2022
|
|
7ENU
 
 | | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, J, Maurya, A, Viswanathan, V, Singh, P.K, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Crystal structure of iron-saturated C-terminal half of lactoferrin produced proteolytically using pepsin at 2.32A resolution
To Be Published
|
|
7WZY
 
 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution | | Descriptor: | ATP phosphoribosyltransferase, FORMIC ACID, GLYCEROL | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.975 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution
To Be Published
|
|
