3KJZ
 
 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
4MAV
 
 | | Crystal structure of signaling protein SPB-40 complexed with 5-hydroxymethyl oxalanetriol at 2.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, GLYCEROL, ... | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-08-17 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of signaling protein SPB-40 complexed with 5-hydroxymethyl oxalanetriol at 2.80 A resolution
To be Published
|
|
4ML4
 
 | | Crystal structure of the complex of signaling glycoprotein from buffalo (SPB-40) with tetrahydropyran at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, TETRAHYDROPYRAN | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of signaling glycoprotein from buffalo (SPB-40) with tetrahydropyran at 2.5 A resolution
To be Published
|
|
1F8M
 
 | | CRYSTAL STRUCTURE OF 3-BROMOPYRUVATE MODIFIED ISOCITRATE LYASE (ICL) FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ISOCITRATE LYASE, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-30 | | Release date: | 2000-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1F61
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE LYASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ISOCITRATE LYASE, MAGNESIUM ION | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K.H, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-19 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1F8I
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE LYASE:NITROPROPIONATE:GLYOXYLATE COMPLEX FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | GLYOXYLIC ACID, ISOCITRATE LYASE, MAGNESIUM ION, ... | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-30 | | Release date: | 2000-12-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1MF4
 
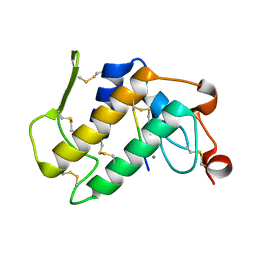 | | Structure-based design of potent and selective inhibitors of phospholipase A2: Crystal structure of the complex formed between phosholipase A2 from Naja Naja sagittifera and a designed peptide inhibitor at 1.9 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2, VAL-ALA-PHE-ARG-SER | | Authors: | Singh, R.K, Vikram, P, Paramsivam, M, Jabeen, T, Sharma, S, Makker, J, Dey, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-08-09 | | Release date: | 2003-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of specific peptide inhibitors for group I phospholipase A2: structure of a complex formed between phospholipase A2 from Naja naja sagittifera (group I) and a designed peptide inhibitor Val-Ala-Phe-Arg-Ser (VAFRS) at 1.9 A resolution reveals unique features
Biochemistry, 42, 2003
|
|
1LN8
 
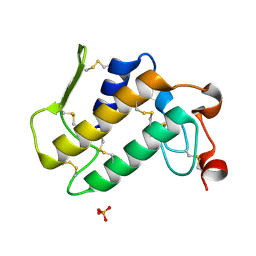 | | Crystal Structure of a New Isoform of Phospholipase A2 from Naja naja sagittifera at 1.6 A Resolution | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Phospholipase A2 | | Authors: | Singh, R.K, Vikram, P, Paramasivam, M, Jabeen, T, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-05-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a New Form of Phospholipase A2 from Naja naja sagittifera at 1.6 A Resolution
to be published
|
|
2PQ2
 
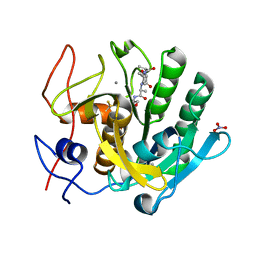 | | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution | | Descriptor: | CALCIUM ION, GALAG peptide, NITRATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, A.K, Singh, N, Sharma, S, Sinha, M, Somvanshi, R.K, Kaur, P, Dey, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution
To be Published
|
|
2Q1P
 
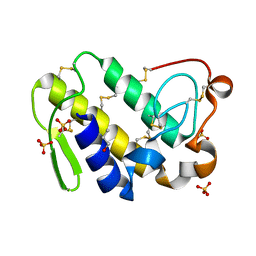 | | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution | | Descriptor: | N-PROPANOL, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Kumar, S, Hariprasad, G, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution
To be Published
|
|
8Y9X
 
 | | Crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, CALCIUM ION, ... | | Authors: | Viswanathan, V, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for the order of preference of inorganic substrates in mammalian heme peroxidases: crystal structure of the complex of lactoperoxidase with four inorganic substrates, SCN, I, Br and Cl
To Be Published
|
|
2PYC
 
 | | Crystal structure of a monomeric phospholipase A2 from Russell's viper at 1.5A resolution | | Descriptor: | ACETATE ION, ACETONITRILE, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2007-05-16 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a monomeric phospholipase A2 from Russell's viper at 1.5A resolution
To be Published
|
|
2PWS
 
 | | Crystal structure of the complex formed between phospholipase A2 and 2-(4-isobutyl-phenyl)-propionic acid at 2.2 A resolution | | Descriptor: | IBUPROFEN, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-05-13 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the complex formed between phospholipase A2 and 2-(4-isobutyl-phenyl)-propionic acid at 2.2 resolution
To be Published
|
|
2QHW
 
 | | Crystal structure of a complex of phospholipase A2 with a gramine derivative at 2.2 resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-07-03 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of a complex of phospholipase A2 with a gramine derivative at 2.2 resolution
TO BE PUBLISHED
|
|
6VTX
 
 | | Crystal structure of human KLF4 zinc finger DNA binding domain in complex with NANOG DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*TP*GP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*CP*AP*CP*CP*CP*CP*CP*T)-3'), Krueppel-like factor 4, ... | | Authors: | Sharma, R, Sharma, S, Choi, K.J, Ferreon, A.C.M, Ferreon, J.C, Sankaran, B, MacKenzie, K.R, Kim, C. | | Deposit date: | 2020-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization.
Nat Commun, 12, 2021
|
|
4ORV
 
 | | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-phenylheptanoic acid, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution
To be Published
|
|
4OUG
 
 | | Crystal structure of the ternary complex of camel peptidoglycan recognition protein, PGRP-S with lipopolysaccharide and palmitic acid at 2.46 A resolution | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-17 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of the ternary complex of camel peptidoglycan recognition protein, PGRP-S with lipopolysaccharide and palmitic acid at 2.46 A resolution
To be Published
|
|
2R2K
 
 | | Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, L(+)-TARTARIC ACID, Peptidoglycan recognition protein | | Authors: | Sharma, P, Jain, R, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-08-26 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of the complex of camel peptidoglycan recognition protein with disaccharide at 3.2A resolution
To be Published
|
|
3T2V
 
 | | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution | | Descriptor: | (2S,3R)-2-hexyl-3-hydroxynonanoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
5H7X
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with 2-hydroxy-1,2,3-propane tricarboxylate at 1.76 A resolution | | Descriptor: | CITRIC ACID, Phosphopantetheine adenylyltransferase | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and binding studies of phosphopantetheine adenylyl transferase from Acinetobacter baumannii.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
2R90
 
 | | Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution | | Descriptor: | Peptidoglycan recognition protein | | Authors: | Sharma, P, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cameline peptidoglycan recognition protein at 2.8A resolution
To be Published
|
|
3CG9
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-L-rhamnopyranose | | Authors: | Sharma, P, Kaur, A, Singh, N, Sharma, S, Bhushan, A, Pathak, K.M.L, Kaur, P, Singh, T.P. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with methyoxane-2,3,4,5-tetrol at 2.9 A resolution
To be Published
|
|
3UIL
 
 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
4FNN
 
 | | Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION | | Descriptor: | Peptidoglycan recognition protein 1, STEARIC ACID | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site.
Arch.Biochem.Biophys., 529, 2013
|
|
3CXA
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Balaji, K, Sharma, P, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution
To be Published
|
|
