6OCK
 
 | | Crystal Structure of Leporine Serum Albumin in Complex with Ketoprofen | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R)-2-{[(2R)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, ... | | Authors: | Zielinski, K, Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-03-24 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigations of stereoselective profen binding by equine and leporine serum albumins.
Chirality, 32, 2020
|
|
6ZTX
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6ZTV
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6SMW
 
 | | A. thaliana serine hydroxymethyltransferase isoform 2 (AtSHMT2) in complex with pemetrexed | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ruszkowski, M, Sekula, B, Dauter, Z. | | Deposit date: | 2019-08-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of methotrexate and pemetrexed action on serine hydroxymethyltransferases revealed using plant models.
Sci Rep, 9, 2019
|
|
6SMN
 
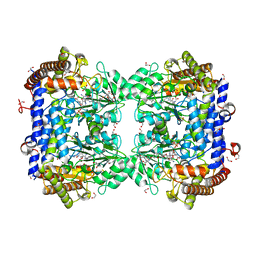 | | A. thaliana serine hydroxymethyltransferase isoform 2 (AtSHMT2) in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, METHOTREXATE, SERINE, ... | | Authors: | Ruszkowski, M, Sekula, B, Dauter, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of methotrexate and pemetrexed action on serine hydroxymethyltransferases revealed using plant models.
Sci Rep, 9, 2019
|
|
6SMR
 
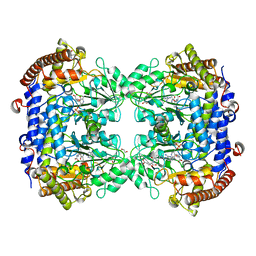 | | A. thaliana serine hydroxymethyltransferase isoform 4 (AtSHMT4) in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, METHOTREXATE, ... | | Authors: | Ruszkowski, M, Sekula, B, Dauter, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis of methotrexate and pemetrexed action on serine hydroxymethyltransferases revealed using plant models.
Sci Rep, 9, 2019
|
|
7Q00
 
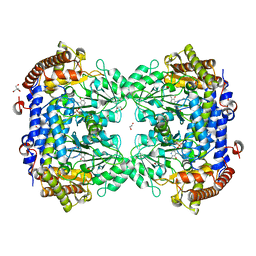 | | Crystal structure of serine hydroxymethyltransferase, isoform 4 from Arabidopsis thaliana (SHM4) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine hydroxymethyltransferase 4 | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7PZZ
 
 | | Crystal structure of serine hydroxymethyltransferase, isoform 2 from Arabidopsis thaliana (SHM2) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Ruszkowski, M, Sekula, B. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
4PO0
 
 | | Crystal Structure of Leporine Serum Albumin in complex with naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Serum albumin | | Authors: | Zielinski, K, Bujacz, A, Sekula, B, Bujacz, G. | | Deposit date: | 2014-02-23 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen.
Proteins, 82, 2014
|
|
7QPE
 
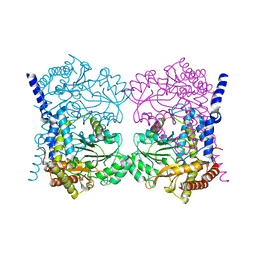 | | Crystal structure of serine hydroxymethyltransferase, isoform 6 from Arabidopsis thaliana (SHM6) | | Descriptor: | NITRATE ION, Serine hydroxymethyltransferase 6 | | Authors: | Ruszkowski, M, Grzechowiak, M, Sekula, B. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
7QX8
 
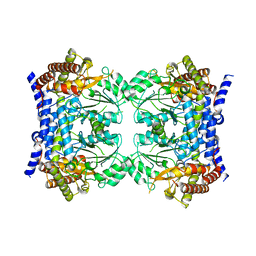 | | Crystal structure of serine hydroxymethyltransferase, isoform 7 from Arabidopsis thaliana (SHM7) | | Descriptor: | Serine hydroxymethyltransferase 7 | | Authors: | Ruszkowski, M, Grzechowiak, M, Sekula, B. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives.
Plant Physiol Biochem., 187, 2022
|
|
4OR0
 
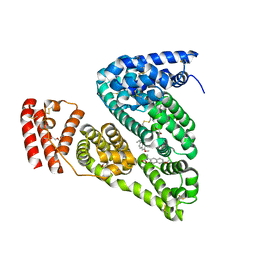 | | Crystal Structure of Bovine Serum Albumin in complex with naproxen | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, DI(HYDROXYETHYL)ETHER, Serum albumin, ... | | Authors: | Zielinski, K, Bujacz, A, Sekula, B, Bujacz, G. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural studies of bovine, equine, and leporine serum albumin complexes with naproxen.
Proteins, 82, 2014
|
|
6YMF
 
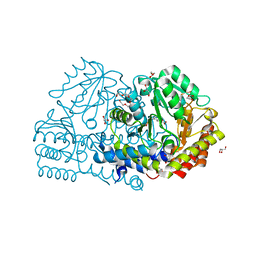 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-Serine external aldimine state | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Serine hydroxymethyltransferase, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YMD
 
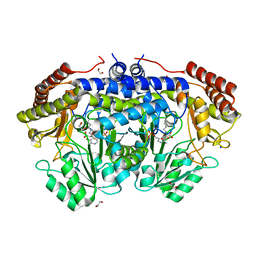 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the covalent complex with malonate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MALONATE ION, ... | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YME
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | Deposit date: | 2020-04-08 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6Z18
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; R32 form | | Descriptor: | RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG | | Authors: | Ruszkowski, M, Sekula, B, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
6CD0
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), PLP-internal aldimine and apo form | | Descriptor: | ACETATE ION, FORMIC ACID, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
6CD1
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), complexes with reaction intermediates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCINE, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
6CCZ
 
 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3) soaked with selenourea | | Descriptor: | ACETATE ION, FORMIC ACID, Serine hydroxymethyltransferase, ... | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
7ALM
 
 | |
