1SMO
 
 | | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47 . | | Descriptor: | L(+)-TARTARIC ACID, triggering receptor expressed on myeloid cells 1 | | Authors: | Kelker, M.S, Foss, T.R, Peti, W, Teyton, L, Kelly, J.W, Wilson, I.A. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47A.
J.Mol.Biol., 342, 2004
|
|
4MOV
 
 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4503 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1RDU
 
 | | NMR STRUCTURE OF A PUTATIVE NIFB PROTEIN FROM THERMOTOGA (TM1290), WHICH BELONGS TO THE DUF35 FAMILY | | Descriptor: | conserved hypothetical protein | | Authors: | Etezady-Esfarjani, T, Herrmann, T, Peti, W, Klock, H.E, Lesley, S.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure Determination of the Hypothetical Protein TM1290 from Thermotoga Maritima using Automated NOESY Analysis.
J.Biomol.NMR, 29, 2004
|
|
4PCV
 
 | | The structure of BdcA (YjgI) from E. coli | | Descriptor: | BdcA (YjgI), PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Page, R, Peti, W, Lord, D. | | Deposit date: | 2014-04-16 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BdcA, a protein important for Escherichia coli biofilm dispersal, is a short-chain dehydrogenase/reductase that binds specifically to NADPH.
Plos One, 9, 2014
|
|
4P9F
 
 | | E. coli McbR/YncC | | Descriptor: | HTH-type transcriptional regulator mcbR | | Authors: | Lord, D.M, Page, R, Peti, W. | | Deposit date: | 2014-04-03 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | McbR/YncC: Implications for the Mechanism of Ligand and DNA Binding by a Bacterial GntR Transcriptional Regulator Involved in Biofilm Formation.
Biochemistry, 53, 2014
|
|
4MOY
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP0
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PHL
 
 | | TbrPDEB1-inhibitor complex | | Descriptor: | 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, Class 1 phosphodiesterase PDEB1, ETHANOL, ... | | Authors: | Choy, M.S, Bland, N, Peti, W, Page, R. | | Deposit date: | 2014-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TbrPDEB1-inhibitor complex
To Be Published
|
|
1T3V
 
 | | The NMR solution structure of TM1816 | | Descriptor: | conserved hypothetical protein | | Authors: | Columbus, L, Peti, W, Herrmann, T, Etazady, T, Klock, H, Lesley, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-04-27 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of the conserved hypothetical protein TM1816 from Thermotoga maritima.
Proteins, 60, 2005
|
|
3E7A
 
 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin Nodularin-R | | Descriptor: | AZIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
3EGH
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1), the PP1 binding and PDZ domains of Spinophilin and the small natural molecular toxin Nodularin-R | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3FMY
 
 | |
3E7B
 
 | | Crystal Structure of Protein Phosphatase-1 Bound to the natural toxin inhibitor Tautomycin | | Descriptor: | (2Z)-2-[(1R)-3-{[(1R,2S,3R,6S,7S,10R)-10-{(2S,3S,6R,8S,9R)-3,9-dimethyl-8-[(3S)-3-methyl-4-oxopentyl]-1,7-dioxaspiro[5.5]undec-2-yl}-3,7-dihydroxy-2-methoxy-6-methyl-1-(1-methylethyl)-5-oxoundecyl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kelker, M.S, Page, R, Peti, W. | | Deposit date: | 2008-08-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors
J.Mol.Biol., 385, 2009
|
|
3EGG
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Spinophilin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3GA8
 
 | | Structure of the N-terminal domain of the E. coli protein MqsA (YgiT/b3021) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, HTH-type transcriptional regulator MqsA (YgiT/B3021), ZINC ION | | Authors: | Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | Deposit date: | 2009-02-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
2L6A
 
 | |
2LLZ
 
 | | GhoS (YjdK) monomer | | Descriptor: | Uncharacterized protein yjdK | | Authors: | Lord, D, Peti, W, Page, R. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS.
Nat.Chem.Biol., 8, 2012
|
|
2LPE
 
 | |
2M3V
 
 | |
2NSV
 
 | |
2NSW
 
 | |
2OXL
 
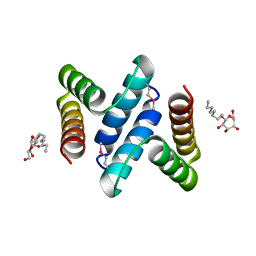 | | Structure and Function of the E. coli Protein YmgB: a Protein Critical for Biofilm Formation and Acid Resistance | | Descriptor: | Hypothetical protein ymgB, octyl beta-D-glucopyranoside | | Authors: | Page, R, Peti, W, Woods, T.K, Palermino, J.M, Doshi, O. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid-resistance.
J.Mol.Biol., 373, 2007
|
|
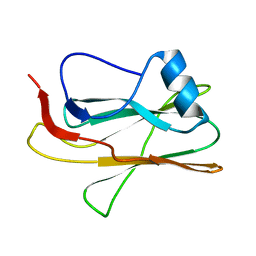
2M83
 
 | |
8DWL
 
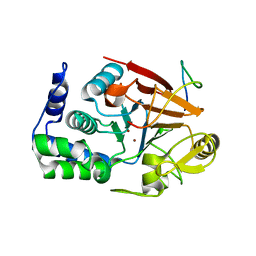 | | Inhibitor-3:PP1 coexpressed complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase PPP1R11, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
8DWK
 
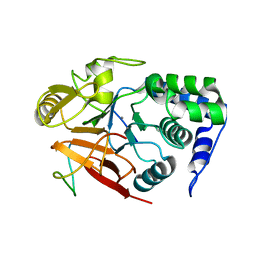 | | Inhibitor-3:PP1 reconstituted complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
