3BQ3
 
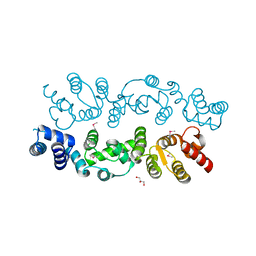 | | Crystal structure of S. cerevisiae Dcn1 | | Descriptor: | Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Chou, Y.C, Sicheri, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-01-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dcn1 Functions as a Scaffold-Type E3 Ligase for Cullin Neddylation.
Mol.Cell, 29, 2008
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
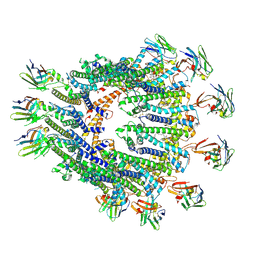
8RMM
 
 | |
8RMK
 
 | |
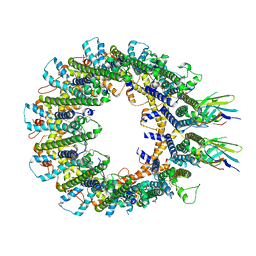
8RML
 
 | |
7K75
 
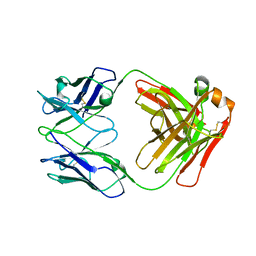 | |
7K76
 
 | |
7ATY
 
 | | Solution NMR structure of the SH3 domain of human Caskin1 | | Descriptor: | Caskin-1 | | Authors: | Toke, O, Koprivanacz, K, Radnai, L, Mero, B, Juhasz, T, Liliom, K, Buday, L. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the SH3 Domain of Human Caskin1 Validates the Lack of a Typical Peptide Binding Groove and Supports a Role in Lipid Mediator Binding.
Cells, 10, 2021
|
|
1PA2
 
 | | ARABIDOPSIS THALIANA PEROXIDASE A2 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PEROXIDASE, ... | | Authors: | Henriksen, A. | | Deposit date: | 1999-05-18 | | Release date: | 2000-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification.
Plant Mol.Biol., 44, 2000
|
|
2KWV
 
 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWU
 
 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | Descriptor: | DNA polymerase iota, Ubiquitin | | Authors: | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2LP9
 
 | | Pseudo-triloop from the sub-genomic promoter of Brome Mosaic Virus | | Descriptor: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*AP*UP*CP*UP*UP*C)-3') | | Authors: | Skov, J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
2LPA
 
 | | Mutant of the sub-genomic promoter from Brome Mosaic Virus | | Descriptor: | RNA (5'-R(*GP*AP*GP*GP*AP*CP*AP*UP*AP*GP*UP*CP*UP*UP*C)-3') | | Authors: | Skov, J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The subgenomic promoter of brome mosaic virus folds into a stem-loop structure capped by a pseudo-triloop that is structurally similar to the triloop of the genomic promoter.
Rna, 18, 2012
|
|
1BKC
 
 | | CATALYTIC DOMAIN OF TNF-ALPHA CONVERTING ENZYME (TACE) | | Descriptor: | N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, TUMOR NECROSIS FACTOR-ALPHA-CONVERTING ENZYME, ZINC ION | | Authors: | Maskos, K, Fernandez-Catalan, C, Bode, W. | | Deposit date: | 1998-04-23 | | Release date: | 1999-06-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
