3U8X
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3SJH
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP-Latrunculin A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-06-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
1B0C
 
 | | EVIDENCE OF A COMMON DECAMER IN THREE CRYSTAL STRUCTURES OF BPTI, CRYSTALLIZED FROM THIOCYANATE, CHLORIDE OR SULFATE | | Descriptor: | PROTEIN (PANCREATIC TRYPSIN INHIBITOR) | | Authors: | Hamiaux, C, Prange, T, Ries-Kautt, M, Ducruix, A, Lafont, S, Astier, J.P, Veesler, S. | | Deposit date: | 1998-11-06 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The BPTI decamer observed in acidic pH crystal forms pre-exists as a stable species in solution.
J.Mol.Biol., 297, 2000
|
|
1BZ5
 
 | | EVIDENCE OF A COMMON DECAMER IN THREE CRYSTAL STRUCTURES OF BPTI, CRYSTALLIZE FROM THIOCYANATE, CHLORIDE OR SULFATE | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Hamiaux, C, Prange, T, Ries-Kautt, M, Ducruix, A, Lafont, S, Astier, J.P, Veesler, S. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The BPTI decamer observed in acidic pH crystal forms pre-exists as a stable species in solution.
J.Mol.Biol., 297, 2000
|
|
5HLP
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | Descriptor: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
5HLN
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH CHIR99021 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHIR99021, Glycogen synthase kinase-3 beta, ... | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
4Z68
 
 | | Hybrid structural analysis of the Arp2/3 regulator Arpin identifies its acidic tail as a primary binding epitope | | Descriptor: | GLU-ILE-ARG-GLU-GLN-GLY-ASP-GLY-ALA-GLU-ASP-GLU, SULFATE ION, Tankyrase-2 | | Authors: | Fetics, S.K, Campanacci, V, Dang, I, Gautreau, A, Cherfils, J. | | Deposit date: | 2015-04-04 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Hybrid Structural Analysis of the Arp2/3 Regulator Arpin Identifies Its Acidic Tail as a Primary Binding Epitope.
Structure, 24, 2016
|
|
4GPK
 
 | | Crystal structure of NprR in complex with its cognate peptide NprX | | Descriptor: | NprR, NprX peptide | | Authors: | Zouhir, S, Guimaraes, B, Perchat, S, Nicaise, M, Lereclus, D, Nessler, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR.
Nucleic Acids Res., 41, 2013
|
|
6GD7
 
 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with PEG | | Descriptor: | Cytochrome c iso-1, GLYCEROL, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6S99
 
 | | Dimethylated fusion protein of RSL and trimeric coiled coil in complex with cucurbit[7]uril | | Descriptor: | 4dzn-RSL,Fucose-binding lectin protein,Fucose-binding lectin protein, beta-D-mannopyranose, cucurbit[7]uril | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2019-07-11 | | Release date: | 2020-08-26 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 2021
|
|
6GD8
 
 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P31 form | | Descriptor: | Cytochrome c iso-1, HEME C, sulfonato-calix[8]arene | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6SU0
 
 | | Crystal structure of dimethylated RSLex in complex with cucurbit[7]uril | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, cucurbit[7]uril, ... | | Authors: | Guagnini, F, Engilberge, S, Crowley, P.B. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineered assembly of a protein-cucurbituril biohybrid.
Chem.Commun.(Camb.), 56, 2020
|
|
3V11
 
 | |
6GD6
 
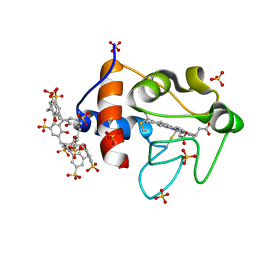 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with ammonium sulfate | | Descriptor: | Cytochrome c iso-1, HEME C, SULFATE ION, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GD9
 
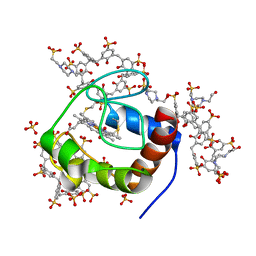 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P43212 form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytochrome c iso-1, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3UKG
 
 | | Crystal structure of Rap1/DNA complex | | Descriptor: | CALCIUM ION, DNA-binding protein RAP1, telomeric DNA | | Authors: | Matot, B, Le Bihan, Y.-V, Gasparini, S, LeDu, M.H. | | Deposit date: | 2011-11-09 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The orientation of the C-terminal domain of the Saccharomyces cerevisiae Rap1 protein is determined by its binding to DNA.
Nucleic Acids Res., 40, 2012
|
|
7KXJ
 
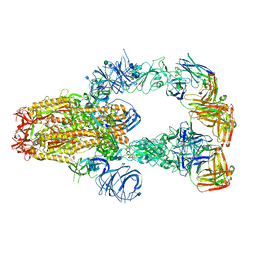 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 3-"up", asymmetric | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KXK
 
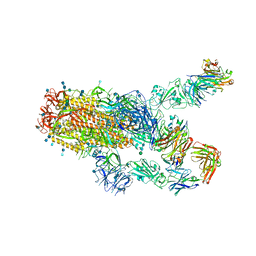 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 2-"up"-1-"down" conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
3F2M
 
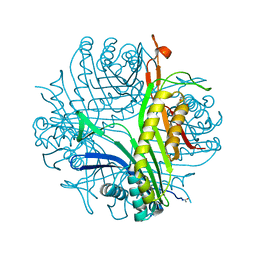 | | Urate oxidase complexed with 8-azaxanthine at 150 MPa | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure.
Biophys.J., 98, 2010
|
|
6STZ
 
 | |
5G2Z
 
 | |
5G30
 
 | |
5G31
 
 | |
8RQP
 
 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), Alginate production protein AlgE, ... | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQR
 
 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (S-form), Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
