2ZVR
 
 | |
3WIC
 
 | | Structure of a substrate/cofactor-unbound glucose dehydrogenase | | Descriptor: | Glucose 1-dehydrogenase, PENTAETHYLENE GLYCOL, S-1,2-PROPANEDIOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1N7K
 
 | | Unique tetrameric structure of deoxyribose phosphate aldolase from Aeropyrum pernix | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Tsuge, H, Sakuraba, H, Shimoya, I, Katunuma, N, Ago, H, Miyano, M, Ohshima, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-15 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The First Crystal Structure of Archaeal Aldolase. UNIQUE TETRAMERIC STRUCTURE of 2-DEOXY-D-RIBOSE-5-PHOSPHATE ALDOLASE FROM THE HYPERTHERMOPHILIC ARCHAEA Aeropyrum pernix.
J.Biol.Chem., 278, 2003
|
|
3WXB
 
 | | Crystal structure of NADPH bound carbonyl reductase from chicken fatty liver | | Descriptor: | 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, Y, Sone, T, Araki, T, Ohshima, T. | | Deposit date: | 2014-07-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A novel NAD(P)H-dependent carbonyl reductase specifically expressed in the thyroidectomized chicken fatty liver: catalytic properties and crystal structure.
Febs J., 282, 2015
|
|
4XB2
 
 | | Hyperthermophilic archaeal homoserine dehydrogenase mutant in complex with NADPH | | Descriptor: | 319aa long hypothetical homoserine dehydrogenase, L-HOMOSERINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
4XB1
 
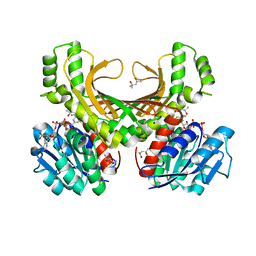 | | Hyperthermophilic archaeal homoserine dehydrogenase in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 319aa long hypothetical homoserine dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
7F76
 
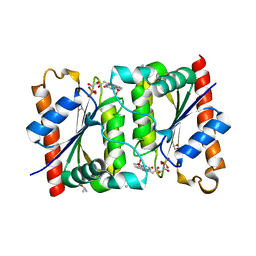 | |
3VPX
 
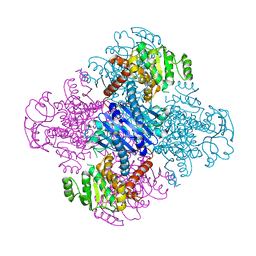 | | Crystal structure of leucine dehydrogenase from a psychrophilic bacterium Sporosarcina psychrophila. | | Descriptor: | Leucine dehydrogenase | | Authors: | Zhao, Y, Wakamatsu, T, Doi, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2012-03-14 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A psychrophilic leucine dehydrogenase from Sporosarcina psychrophila: Purification, characterization, gene sequencing and crystal structure analysis
J.MOL.CATAL., B ENZYM., 83, 2012
|
|
3VQR
 
 | | Structure of a dye-linked L-proline dehydrogenase mutant from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
2YVS
 
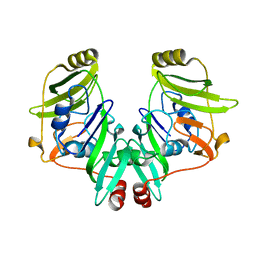 | |
2ZC0
 
 | | Crystal structure of an archaeal alanine:glyoxylate aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Alanine glyoxylate transaminase, IMIDAZOLE, ... | | Authors: | Sakuraba, H, Yoneda, K, Tsuge, H, Ohshima, T. | | Deposit date: | 2007-10-31 | | Release date: | 2008-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an archaeal alanine:glyoxylate aminotransferase
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3VTF
 
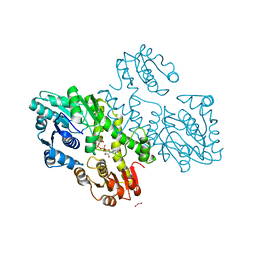 | | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a UDP-glucose dehydrogenase from the hyperthermophilic archaeon Pyrobaculum islandicum.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3W6Z
 
 | |
1WY6
 
 | | Crystal Structure of Hypothetical Protein [ST1625p] from Hyperthermophilic Archaeon Sulfolobus tokodaii | | Descriptor: | hypothetical protein ST1625 | | Authors: | Yoneda, K, Sakuraba, H, Tsuge, H, Katunuma, N, Kuramitsu, S, Kawabata, T, Ohshima, T. | | Deposit date: | 2005-02-07 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The first crystal structure of an archaeal helical repeat protein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3WID
 
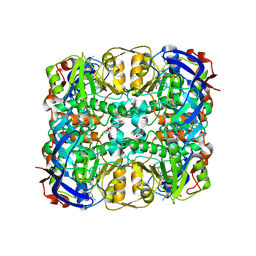 | | Structure of a glucose dehydrogenase T277F mutant in complex with NADP | | Descriptor: | Glucose 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WIE
 
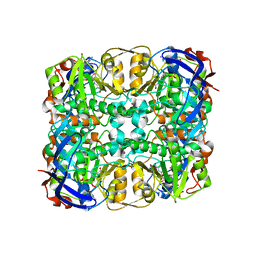 | | Structure of a glucose dehydrogenase T277F mutant in complex with D-glucose and NAADP | | Descriptor: | Glucose 1-dehydrogenase, ZINC ION, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WQO
 
 | | Crystal structure of D-tagatose 3-epimerase-like protein | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein MJ1311 | | Authors: | Uechi, K, Takata, G, Yoneda, K, Ohshima, T, Sakuraba, H. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of D-tagatose 3-epimerase-like protein from Methanocaldococcus jannaschii
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1VCV
 
 | |
2E5V
 
 | | Crystal structure of L-Aspartate Oxidase from hyperthermophilic archaeon Sulfolobus tokodaii | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, L-aspartate oxidase | | Authors: | Yoneda, K, Sakuraba, H, Asai, I, Tsuge, H, Katunuma, N, Ohshima, T. | | Deposit date: | 2006-12-25 | | Release date: | 2008-01-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of l-aspartate oxidase from the hyperthermophilic archaeon Sulfolobus tokodaii
Biochim.Biophys.Acta, 1784, 2008
|
|
1Y56
 
 | | Crystal structure of L-proline dehydrogenase from P.horikoshii | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tsuge, H, Kawakami, R, Sakuraba, H, Ago, H, Miyano, M, Aki, K, Katunuma, N, Ohshima, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure of a novel FAD-, FMN-, and ATP-containing L-proline dehydrogenase complex from Pyrococcus horikoshii
J.Biol.Chem., 280, 2005
|
|
2DCH
 
 | | Crystal structure of archaeal intron-encoded homing endonuclease I-Tsp061I | | Descriptor: | CHLORIDE ION, SULFATE ION, putative homing endonuclease | | Authors: | Nakayama, H, Tsuge, H, Shimamura, T, Miyano, M, Nomura, N, Sako, Y. | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of a hyperthermophilic archaeal homing endonuclease, I-Tsp061I: contribution of cross-domain polar networks to thermostability.
J.Mol.Biol., 365, 2007
|
|
5AVO
 
 | |
7F4B
 
 | | The crystal structure of the immature apo-enzyme of homoserine dehydrogenase from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | MAGNESIUM ION, homoserine dehydrogenase | | Authors: | Kurihara, E, Kubota, T, Watanabe, K, Ogata, K, Kaneko, R, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
7F4C
 
 | | The crystal structure of the immature holo-enzyme of homoserine dehydrogenase complexed with NADP and 1,4-butandiol from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | 1,4-BUTANEDIOL, Homoserine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogata, K, Kaneko, R, Kubota, T, Watanabe, K, Kurihara, E, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
5X9D
 
 | | Crystal structure of homoserine dehydrogenase in complex with L-cysteine and NAD | | Descriptor: | (2R)-3-[[(4S)-3-aminocarbonyl-1-[(2R,3R,4S,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4H-pyridin-4-yl]sulfanyl]-2-azanyl-propanoic acid, Homoserine dehydrogenase, L(+)-TARTARIC ACID | | Authors: | Goto, M, Ogata, K, Kaneko, R, Yoshimune, K. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of homoserine dehydrogenase by formation of a cysteine-NAD covalent complex
Sci Rep, 8, 2018
|
|
