8OG4
 
 | | Exostosin-like 3 UDP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, MANGANESE (II) ION, ... | | Authors: | Sammon, D, Hohenester, E. | | Deposit date: | 2023-03-18 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of decision-making in glycosaminoglycan biosynthesis.
Nat Commun, 14, 2023
|
|
6FW9
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 6-Fluoro-L-Tryptophan | | Descriptor: | 6-FLUORO-L-TRYPTOPHAN, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
6FWA
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 7-Methyl-L-Tryptophan | | Descriptor: | 7-Methyl-L-Tryptophan, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.846 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
6FW8
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with 5-Methyl-L-Tryptophan | | Descriptor: | 5-methyl-L-tryptophan, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA, ... | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2018-03-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A GenoChemetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2019
|
|
7QLL
 
 | |
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLK
 
 | |
7QLJ
 
 | |
7QLN
 
 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLO
 
 | | rsKiiro pump dump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLM
 
 | |
7NJZ
 
 | | X-ray crystallography study of RoAb13 which binds to PIYDIN, a part of the CCR5 N terminal domain | | Descriptor: | Antibody RoAb13 Heavy Chain, Antibody RoAb13 Light Chain, Region from C-C chemokine receptor type 5 N-terminal domain | | Authors: | Helliwell, J.R, Chayen, N, Saridakis, E, Govada, L. | | Deposit date: | 2021-02-17 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5.
Iucrj, 8, 2021
|
|
8C4A
 
 | |
7WYO
 
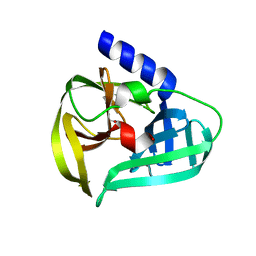 | | Structure of the EV71 3Cpro with 338 inhibitor | | Descriptor: | 3C protein, N-methyl-N-(4,5,6,7-tetrahydro-1,3-benzothiazol-2-ylmethyl)prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYL
 
 | | Structure of the EV71 3Cpro with 337 inhibitor | | Descriptor: | 3C protein, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYM
 
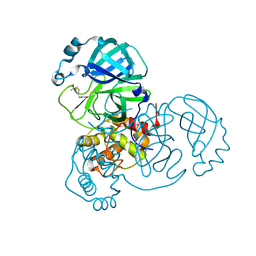 | | Structure of the SARS-COV-2 main protease with 337 inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYP
 
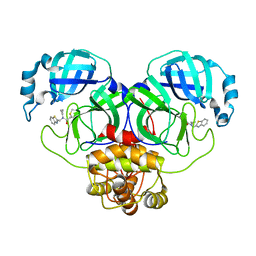 | | Structure of the SARS-COV-2 main protease with EN102 inhibitor | | Descriptor: | 3C-like proteinase, N-(1,3-benzothiazol-2-ylmethyl)-N-cyclopropyl-prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7OML
 
 | |
7OLH
 
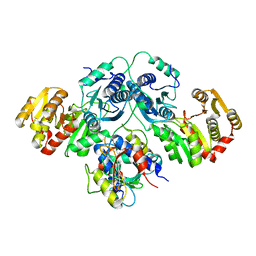 | |
7OJR
 
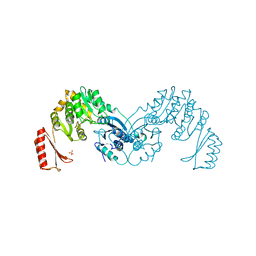 | |
7OJS
 
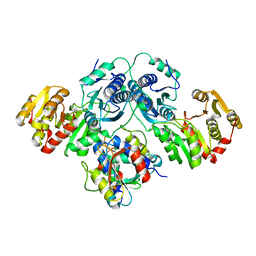 | |
6I3V
 
 | | x-ray structure of the human mitochondrial PRELID1 in complex with TRIAP1 | | Descriptor: | CHLORIDE ION, MYRISTIC ACID, PRELI domain-containing protein 1, ... | | Authors: | Berry, J.L, Miliara, X, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-07 | | Release date: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I3Y
 
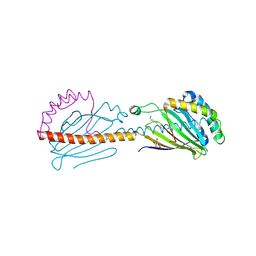 | | Crystal structure of the human mitochondrial PRELID1K58V-TRIAP1 complex with PS | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, PRELI domain-containing protein 1, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I4Y
 
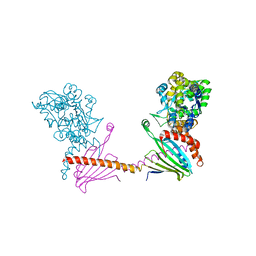 | | X-ray structure of the human mitochondrial PRELID3b-TRIAP1 complex | | Descriptor: | Maltose transport system, substrate-binding protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3B, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
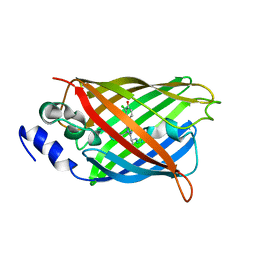
8A6G
 
 | |
