8GSU
 
 | | Crystal structure of human cardiac alpha actin (WT_ADP-Pi) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT4
 
 | | Crystal structure of human cardiac alpha actin Q137A mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT1
 
 | | Crystal structure of human cardiac alpha actin A108G mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GSW
 
 | | Crystal structure of human cardiac alpha actin A108G mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT2
 
 | | Crystal structure of human cardiac alpha actin P109A mutant (AMPPNP state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, Actin, alpha cardiac muscle 1, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT5
 
 | | Crystal structure of human cardiac alpha actin Q137A mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8GT3
 
 | | Crystal structure of human cardiac alpha actin P109A mutant (ADP-Pi state) in complex with fragmin F1 domain | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Iwasa, M, Oda, T, Takeda, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenic analysis of actin reveals the mechanism of His161 flipping that triggers ATP hydrolysis.
Front Cell Dev Biol, 11, 2023
|
|
8H4V
 
 | | Mincle CRD complex with PGL trisaccharide | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol-(1-4)-6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-3-O-methyl-alpha-L-rhamnopyranose, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
8HB5
 
 | | Crystal structure of Mincle in complex with HD-275 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
7W51
 
 | |
7W4Z
 
 | |
7W50
 
 | |
7W52
 
 | |
7YNE
 
 | |
1PGV
 
 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
7EA6
 
 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
8I09
 
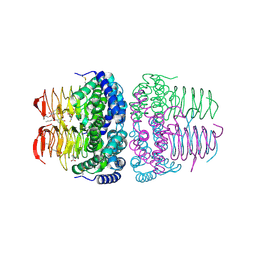 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with butyl gallate | | Descriptor: | CYSTEINE, PHOSPHATE ION, Serine acetyltransferase, ... | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I04
 
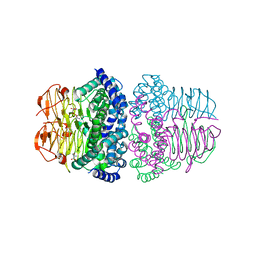 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I06
 
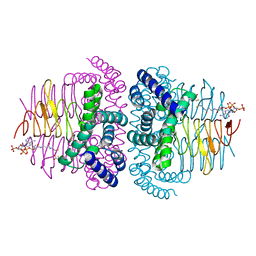 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with CoA | | Descriptor: | COENZYME A, CYSTEINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
5YU8
 
 | | Cofilin decorated actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Tanaka, K, Narita, A. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for cofilin binding and actin filament disassembly
Nat Commun, 9, 2018
|
|
7DF7
 
 | |
7DSA
 
 | |
7DS8
 
 | |
7DS4
 
 | |
7DS3
 
 | |
