8DN2
 
 | |
8DN5
 
 | |
8DN3
 
 | |
8DN4
 
 | | Cryo-EM structure of human Glycine Receptor alpha-1 beta heteromer, glycine-bound state3(desensitized state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-1, Glycine receptor subunit beta,Green fluorescent protein,Glycine receptor beta, ... | | Authors: | Liu, X, Wang, W. | | Deposit date: | 2022-07-10 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric gating of a human hetero-pentameric glycine receptor.
Nat Commun, 14, 2023
|
|
6N48
 
 | | Structure of beta2 adrenergic receptor bound to BI167107, Nanobody 6B9, and a positive allosteric modulator | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Liu, X, Masoudi, A, Kahsai, A.W, Huang, L.Y, Pani, B, Hirata, K, Ahn, S, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2018-11-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of beta2AR regulation by an intracellular positive allosteric modulator.
Science, 364, 2019
|
|
6NWL
 
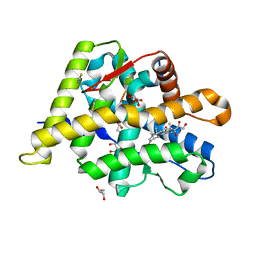 | | Structure of the Ancestral Glucocorticoid Receptor 2 ligand binding domain in complex with hydrocortisone and PGC1a coregulator fragment | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Liu, X, Ortlund, E.A. | | Deposit date: | 2019-02-06 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | First High-Resolution Crystal Structures of the Glucocorticoid Receptor Ligand-Binding Domain-Peroxisome Proliferator-ActivatedgammaCoactivator 1-alphaComplex with Endogenous and Synthetic Glucocorticoids.
Mol.Pharmacol., 96, 2019
|
|
6NWK
 
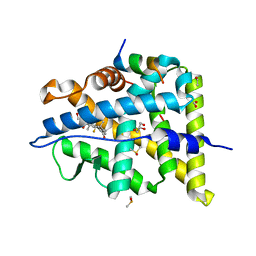 | |
8GUD
 
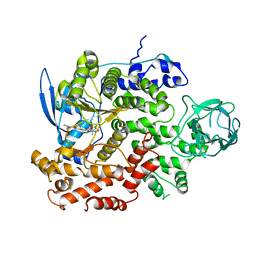 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E545K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUB
 
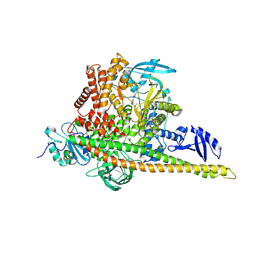 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GUA
 
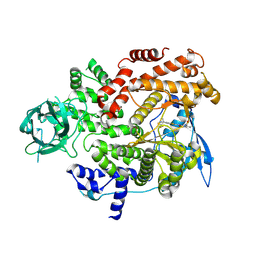 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E542K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5F72
 
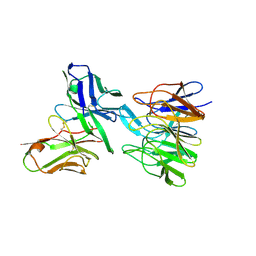 | | De novo design and crystallographic validation of antibodies targeting a pre-selected epitope | | Descriptor: | Kelch-like ECH-associated protein 1, Single chain Fv from a Fab | | Authors: | Liu, X, Taylor, R.D, Griffin, L, Coker, S, Adams, R, Ceska, T, Shi, J, Lawson, A.D.G, Baker, T. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo design and crystallographic validation of antibodies targeting a pre-selected epitope
To Be Published
|
|
7SPC
 
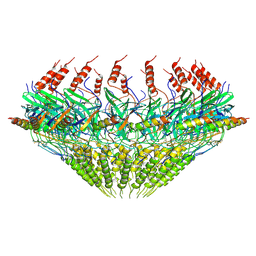 | | Models for C17 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208). | | Descriptor: | TraB, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPJ
 
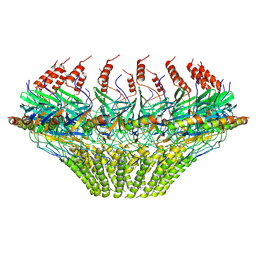 | | Models for C17 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208 | | Descriptor: | TraB, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPI
 
 | | Models for C13 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208 | | Descriptor: | TraB, TraK, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPK
 
 | | Models for C16 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208 | | Descriptor: | TraB, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPB
 
 | | Models for C13 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208). | | Descriptor: | TraB, TraK, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
3O72
 
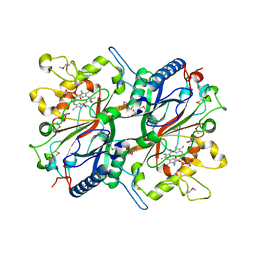 | | Crystal structure of EfeB in complex with heme | | Descriptor: | OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, Redox component of a tripartite ferrous iron transporter | | Authors: | Liu, X, Du, Q, Wang, Z, Zhu, D, Huang, Y, Li, N, Xu, S, Gu, L. | | Deposit date: | 2010-07-30 | | Release date: | 2011-03-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes
J.Biol.Chem., 286, 2011
|
|
6AAX
 
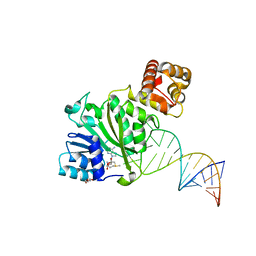 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
7U1A
 
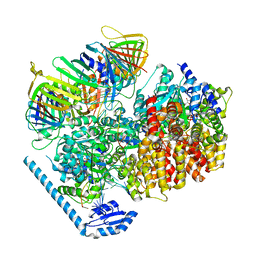 | | RFC:PCNA bound to dsDNA with a ssDNA gap of six nucleotides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
6AJK
 
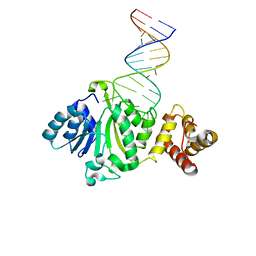 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AAS
 
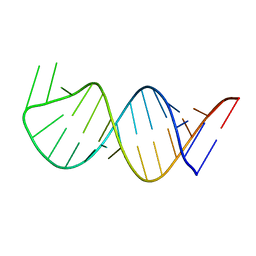 | | Solution Structure for helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (28-MER) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
3TGV
 
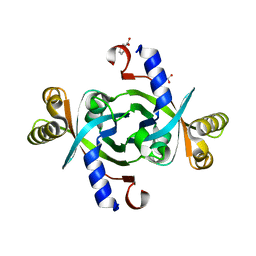 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
4IGK
 
 | |
4IFI
 
 | |
