6JC5
 
 | |
6C7T
 
 | |
6CAI
 
 | |
6C8B
 
 | |
6C7H
 
 | |
6C7M
 
 | |
6C7V
 
 | |
7QLL
 
 | |
7QLK
 
 | |
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLJ
 
 | |
7QLM
 
 | |
7QLN
 
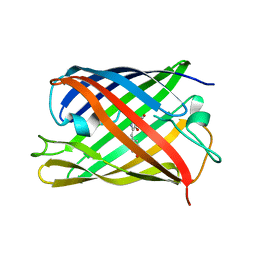 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLO
 
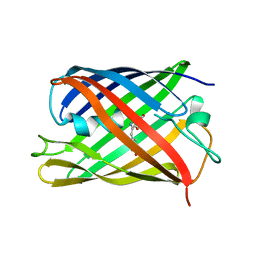 | | rsKiiro pump dump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
8K5R
 
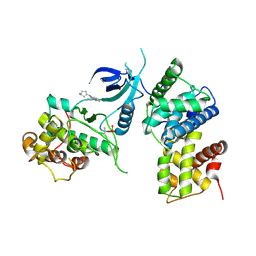 | | CDK9/cyclin T1 in complex with KB-0742 | | Descriptor: | (1S,3S)-N3-(5-pentan-3-ylpyrazolo[1,5-a]pyrimidin-7-yl)cyclopentane-1,3-diamine, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Zhou, M, Li, H, Gao, H, Trotter, B.W, Freeman, D. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Discovery of KB-0742, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of CDK9 for MYC-Dependent Cancers.
J.Med.Chem., 66, 2023
|
|
7MH4
 
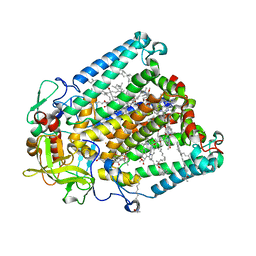 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-bromotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH9
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-nitrotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH5
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-iodotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH8
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-methyltyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MH3
 
 | | Crystal structure of R. sphaeroides Photosynthetic Reaction Center variant; Y(M210)3-chlorotyrosine | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Mathews, I, Weaver, J.B, Boxer, S.G. | | Deposit date: | 2021-04-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1MOZ
 
 | | ADP-ribosylation factor-like 1 (ARL1) from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-like protein 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Amor, J.C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structures of Yeast ARF2 and ARL1:
DISTINCT ROLES FOR THE N TERMINUS IN THE STRUCTURE
AND FUNCTION OF ARF FAMILY GTPases
J.Biol.Chem., 276, 2001
|
|
7MHA
 
 | |
3VHN
 
 | | Y61G mutant of Cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
3VHP
 
 | | The insertion mutant Y61GG of Tm Cel12A | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
3VHO
 
 | | Y61-gg insertion mutant of Tm-Cellulase 12A | | Descriptor: | Endo-1,4-beta-glucanase | | Authors: | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
