3EQA
 
 | | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glucoamylase, ... | | Authors: | Lee, J, Paetzel, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the catalytic domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4MVD
 
 | | Crystal Structure of a Mammalian Cytidylyltransferase | | Descriptor: | Choline-phosphate cytidylyltransferase A, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Lee, J, Cornell, R.B. | | Deposit date: | 2013-09-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Structural Basis for Autoinhibition of CTP:Phosphocholine Cytidylyltransferase (CCT), the Regulatory Enzyme in Phosphatidylcholine Synthesis, by Its Membrane-binding Amphipathic Helix.
J.Biol.Chem., 289, 2014
|
|
4MVC
 
 | | Crystal Structure of a Mammalian Cytidylyltransferase | | Descriptor: | Choline-phosphate cytidylyltransferase A, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Lee, J, Cornell, R.B. | | Deposit date: | 2013-09-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Autoinhibition of CTP:Phosphocholine Cytidylyltransferase (CCT), the Regulatory Enzyme in Phosphatidylcholine Synthesis, by Its Membrane-binding Amphipathic Helix.
J.Biol.Chem., 289, 2014
|
|
4QXL
 
 | | Crystal Structure of FLHE | | Descriptor: | Flagellar protein flhE | | Authors: | Lee, J, Monzingo, A.F, Keatinge-Clay, A.T, Harshey, R.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-01-14 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Structure of Salmonella FlhE, Conserved Member of a Flagellar Type III Secretion Operon.
J.Mol.Biol., 427, 2015
|
|
3O3Q
 
 | |
3PD7
 
 | |
3O49
 
 | |
3OL0
 
 | |
3OGF
 
 | |
7DOG
 
 | |
2LXJ
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp with dT7 | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2LXK
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2PLK
 
 | |
2PLJ
 
 | |
7JP1
 
 | |
7JOY
 
 | |
7KHP
 
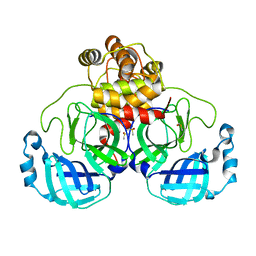 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
8IKQ
 
 | |
1T47
 
 | | Structure of fe2-HPPD bound to NTBC | | Descriptor: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Brownlee, J, Johnson-Winters, K, Harrison, D.H.T, Moran, G.R. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ferrous Form of (4-Hydroxyphenyl)pyruvate Dioxygenase from Streptomyces avermitilis in Complex with the Therapeutic Herbicide, NTBC
Biochemistry, 43, 2004
|
|
8X3N
 
 | |
8X40
 
 | |
2PNM
 
 | |
6ZIF
 
 | |
2F0Y
 
 | |
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | Descriptor: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2005-12-23 | | Release date: | 2007-01-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
