6M04
 
 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
5ZSU
 
 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6LBH
 
 | |
8H1J
 
 | | Cryo-EM structure of the TnpB-omegaRNA-target DNA ternary complex | | Descriptor: | Non-target strand, RNA-guided DNA endonuclease TnpB, Target strand, ... | | Authors: | Nakagawa, R, Hirano, H, Omura, S, Nureki, O. | | Deposit date: | 2022-10-03 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the transposon-associated TnpB enzyme.
Nature, 616, 2023
|
|
7VPK
 
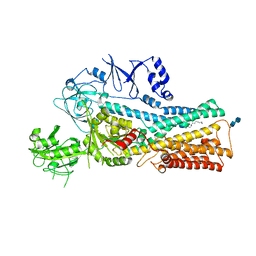 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPI
 
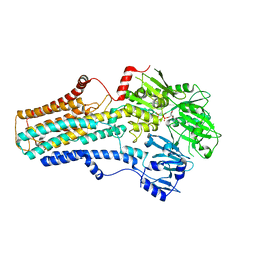 | | Cryo-EM structure of the human ATP13A2 (E1-ATP state) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Polyamine-transporting ATPase 13A2 | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPL
 
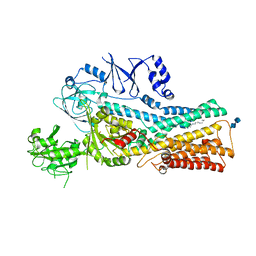 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2Pi state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPJ
 
 | | Cryo-EM structure of the human ATP13A2 (E1P-ADP state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7W9W
 
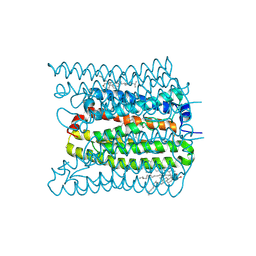 | | 2.02 angstrom cryo-EM structure of the pump-like channelrhodopsin ChRmine | | Descriptor: | CHOLESTEROL, ChRmine, PALMITIC ACID, ... | | Authors: | Kishi, K.E, Kim, Y, Fukuda, M, Yamashita, K, Deisseroth, K, Kato, H.E. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural basis for channel conduction in the pump-like channelrhodopsin ChRmine.
Cell, 185, 2022
|
|
7V73
 
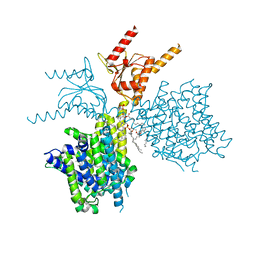 | | Thermostabilized human prestin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V74
 
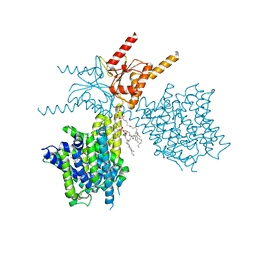 | | Thermostabilized human prestin in complex with sulfate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, SULFATE ION, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V75
 
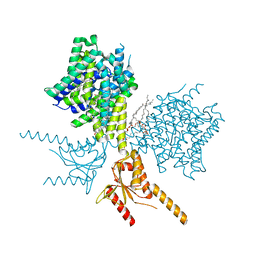 | | Thermostabilized human prestin in complex with salicylate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-HYDROXYBENZOIC ACID, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7E7Z
 
 | |
7E83
 
 | |
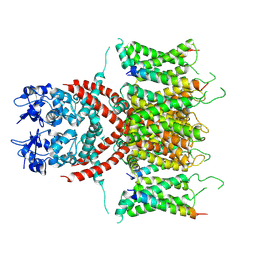
7E87
 
 | |
7E8H
 
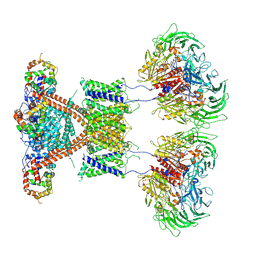 | | CryoEM structure of human Kv4.2-DPP6S-KChIP1 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E84
 
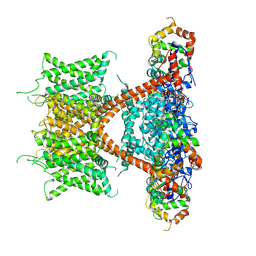 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-02-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7E8E
 
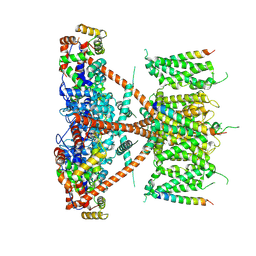 | |
7E8G
 
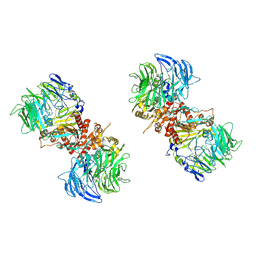 | |
7E89
 
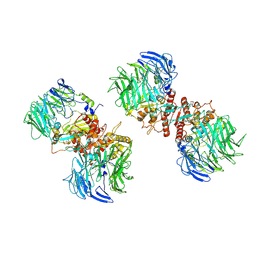 | |
7E8B
 
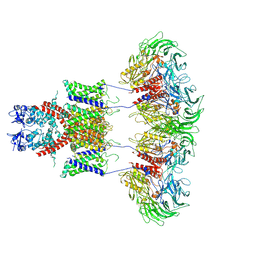 | | CryoEM structure of human Kv4.2-DPP6S complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7F3F
 
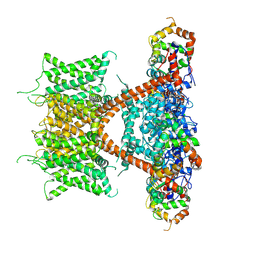 | | CryoEM structure of human Kv4.2-KChIP1 complex | | Descriptor: | Isoform 2 of Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7F0J
 
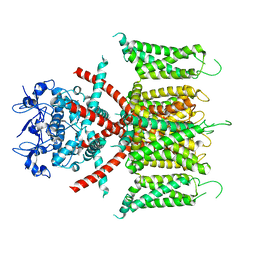 | | CryoEM structure of human Kv4.2 | | Descriptor: | Potassium voltage-gated channel subfamily D member 2, ZINC ION | | Authors: | Kise, Y, Nureki, O. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of gating modulation of Kv4 channel complexes.
Nature, 599, 2021
|
|
7F47
 
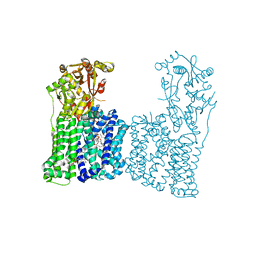 | | Cryo-EM structure of Rhizobium etli MprF | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Hypothetical conserved protein, [(2R)-1-[[(2R)-3-[(2S)-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (E)-octadec-9-enoate | | Authors: | Nishimura, M, Hirano, H, Kobayashi, K, Gill, C.P, Phan, C.N.K, Kise, Y, Kusakizako, T, Yamashita, K, Ito, Y, Roy, H, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of Rhizobium etli MprF
To Be Published
|
|
