1O35
 
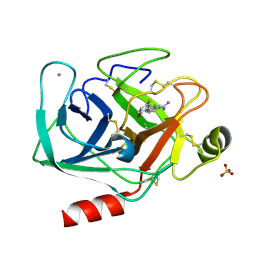 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-4-FLUOROBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
1O3C
 
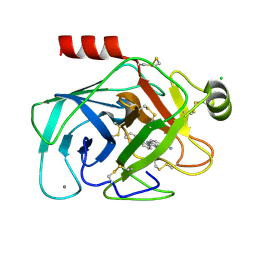 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-1,1'-BIPHENYL-2-OLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
1O3K
 
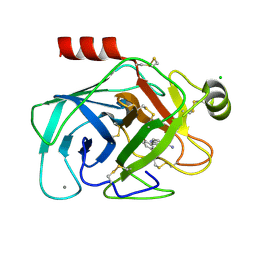 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-6-BROMO-4-METHYLBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W.D, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, V.W, Hui, H.C, Breitenbucher, J.G, Allen, D, Janc, J.W. | | Deposit date: | 2003-03-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Elaborate manifold of short hydrogen bond arrays mediating binding of active site-directed serine protease
inhibitors.
J.Mol.Biol., 329, 2003
|
|
7N1B
 
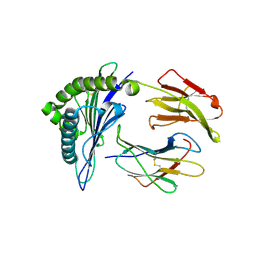 | | SARS-CoV-2 RLQ peptide binds to HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1A
 
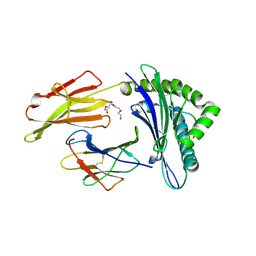 | | SARS-CoV-2 YLQ peptide binds to HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1C
 
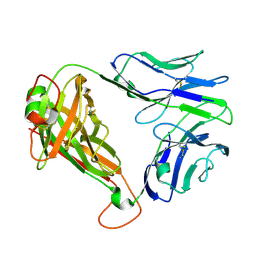 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 | | Descriptor: | pRLQ3 T cell receptor alpha chain, pRLQ3 T cell receptor beta chain | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1D
 
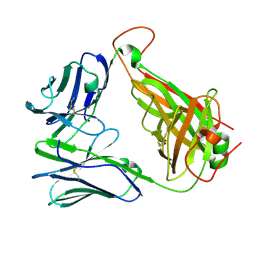 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 | | Descriptor: | pYLQ7 T cell receptor alpha chain, pYLQ7 T cell receptor beta chain | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1F
 
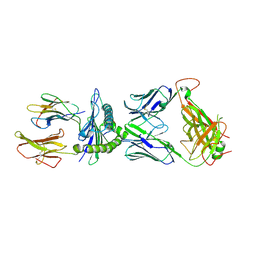 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 binds to YLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1E
 
 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 binds to RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
6NO9
 
 | | PIM1 in complex with Cpd16 (5-amino-N-(5-((4R,5R)-4-amino-5-fluoroazepan-1-yl)-1-methyl-1H-pyrazol-4-yl)-2-(2,6-difluorophenyl)thiazole-4-carboxamide) | | Descriptor: | 5-amino-N-{5-[(4R,5R)-4-amino-5-fluoroazepan-1-yl]-1-methyl-1H-pyrazol-4-yl}-2-(2,6-difluorophenyl)-1,3-thiazole-4-carboxamide, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Murray, J.M, Noland, C. | | Deposit date: | 2019-01-15 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Optimization of Pan-Pim Kinase Activity and Oral Bioavailability Leading to Diaminopyrazole (GDC-0339) for the Treatment of Multiple Myeloma.
J. Med. Chem., 62, 2019
|
|
6WKM
 
 | |
8SO3
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
8SR0
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
4HLE
 
 | | Compound 21 (1-alkyl-substituted 1,2,4-triazoles) | | Descriptor: | 2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-4,5-dihydrothieno[3,2-d][1]benzoxepine-8-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Rouge, L, Wu, P. | | Deposit date: | 2012-10-16 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Cis-Amide isosteric replacement in thienobenzoxepin inhibitors of PI3-kinase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J6I
 
 | | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform | | Descriptor: | 2-methyl-1-(4-{2-[1-(2,2,2-trifluoroethyl)-1H-1,2,4-triazol-5-yl]-4,5-dihydro[1]benzoxepino[5,4-d][1,3]thiazol-8-yl}-1H-pyrazol-1-yl)propan-2-ol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Rouge, L, Wu, P. | | Deposit date: | 2013-02-11 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7SZR
 
 | | NIK bound to inhibitor G02792917 | | Descriptor: | 1-(3-{[(1R,4R,5S)-4-hydroxy-2-methyl-3-oxo-2-azabicyclo[3.1.0]hexan-4-yl]ethynyl}phenyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Liau, N.P.D, Hymowitz, S.G. | | Deposit date: | 2021-11-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Filling a nick in NIK: Extending the half-life of a NIK inhibitor through structure-based drug design.
Bioorg.Med.Chem.Lett., 89, 2023
|
|
5V82
 
 | | PIM1 kinase in complex with Cpd17 (1-(6-(4,4-difluoropiperidin-3-yl)pyridin-2-yl)-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine) | | Descriptor: | 1-{6-[(3R)-4,4-difluoropiperidin-3-yl]pyridin-2-yl}-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Discovery of 5-Azaindazole (GNE-955) as a Potent Pan-Pim Inhibitor with Optimized Bioavailability.
J. Med. Chem., 60, 2017
|
|
5V80
 
 | |
6O9D
 
 | | Structure of the IRAK4 kinase domain with compound 5 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{7-[4-(aminomethyl)piperidin-1-yl]quinolin-6-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6O8U
 
 | | Crystal structure of IRAK4 in complex with compound 23 | | Descriptor: | GLYCEROL, Interleukin-1 receptor-associated kinase 4, N-[2,2-dimethyl-6-(morpholin-4-yl)-2,3-dihydro-1-benzofuran-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, ... | | Authors: | Kiefer, J.R, Yu, C, Drobnick, J, Bryan, M.C, Lupardus, P.J. | | Deposit date: | 2019-03-12 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6O95
 
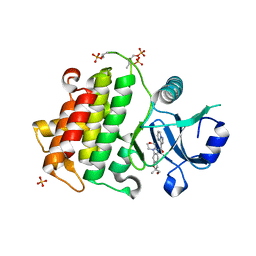 | | Structure of the IRAK4 kinase domain with compound 41 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-(hydroxymethyl)-2-methyl-6-(morpholin-4-yl)-2,3-dihydro-1-benzofuran-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6OQI
 
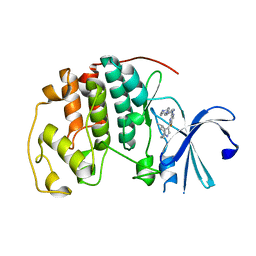 | | CDK2 in complex with Cpd14 (5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine) | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4S)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OQO
 
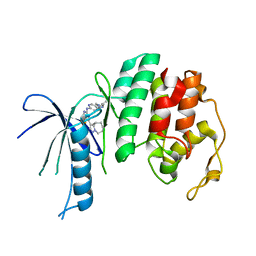 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7R6J
 
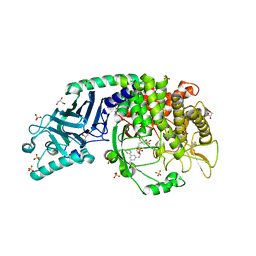 | | Co-crystal structure of Chaetomium glucosidase with compound 1 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[3-methyl-5-(pyrimidin-2-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
7REV
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 3 | | Descriptor: | (2R,3R,4R,5S)-1-[(4-{[4-(furan-2-yl)-2-methylanilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
