2GT7
 
 | | Crystal structure of SARS coronavirus main peptidase at pH 6.0 in the space group P21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2A4O
 
 | | Dual modes of modification of Hepatitis A virus 3C protease by a serine derived beta-lactone: selective crytstallization and high resolution structure of the His102 adduct | | Descriptor: | ACETYL GROUP, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PHENYLALANINE AMIDE, ... | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
1CQJ
 
 | | CRYSTAL STRUCTURE OF DEPHOSPHORYLATED E. COLI SUCCINYL-COA SYNTHETASE | | Descriptor: | COENZYME A, PHOSPHATE ION, SUCCINYL-COA SYNTHETASE ALPHA CHAIN, ... | | Authors: | Joyce, M.A, Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 1999-08-06 | | Release date: | 2000-01-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography.
Biochemistry, 39, 2000
|
|
1CQI
 
 | | Crystal Structure of the Complex of ADP and MG2+ with Dephosphorylated E. Coli Succinyl-CoA Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Joyce, M.A, Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 1999-08-06 | | Release date: | 2000-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography.
Biochemistry, 39, 2000
|
|
2SGP
 
 | | PRO 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | OVOMUCOID INHIBITOR, PHOSPHATE ION, PROTEINASE B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
3BUE
 
 | | Crystal structure of the C-terminal domain hexamer of ArgR from Mycobacterium tuberculosis | | Descriptor: | Arginine repressor ArgR | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-01-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2SEC
 
 | |
2SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water molecules participate in proteinase-inhibitor interactions: crystal structures of Leu18, Ala18, and Gly18 variants of turkey ovomucoid inhibitor third domain complexed with Streptomyces griseus proteinase B.
Protein Sci., 4, 1995
|
|
1BXO
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
1BXQ
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) COMPLEX WITH PHOSPHONATE INHIBITOR. | | Descriptor: | 2-[(1R)-1-(N-(3-METHYLBUTANOYL)-L-VALYL-L-ASPARAGINYL)-AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-PHENYLPROPANOIC ACID METHYLESTER, ACETATE ION, GLYCEROL, ... | | Authors: | Parrish, J.C, Khan, A.R, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
2D3Z
 
 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3U
 
 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2DJ5
 
 | | Crystal Structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Cherney, M.M, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Vitamin B(12) Biosynthetic Cobaltochelatase, CbiX (S), from Archaeoglobus Fulgidus
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
2D41
 
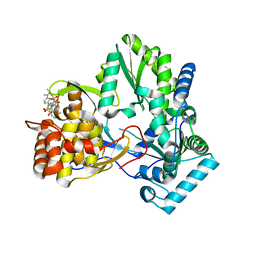 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | Descriptor: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-05 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2CXV
 
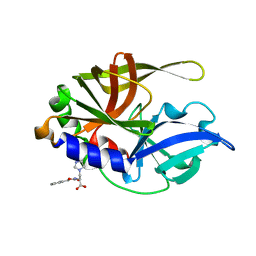 | | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-Derived betaLactone: Selective Crystallization and High-resolution Structure of the His-102 Adduct | | Descriptor: | N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Probable protein P3C | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-07-01 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
2NR9
 
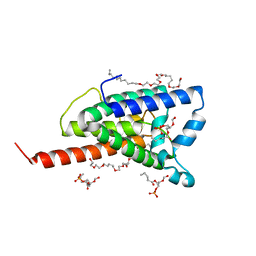 | | Crystal structure of GlpG, Rhomboid Peptidase from Haemophilus influenzae | | Descriptor: | (R)-2-(FORMYLOXY)-3-(PHOSPHONOOXY)PROPYL PENTANOATE, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, Protein glpG homolog | | Authors: | Lemieux, M.J, Fischer, S.J, Cherney, M.M, Bateman, K.S, James, M.N.G. | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NU0
 
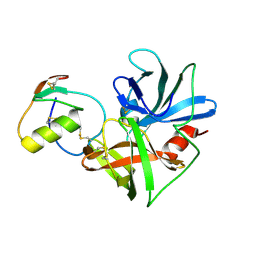 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I
To be Published
|
|
2NU1
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I
To be Published
|
|
2NU4
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2O2H
 
 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane | | Descriptor: | 1,2-DICHLOROETHANE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane
TO BE PUBLISHED
|
|
2NU3
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
2O2I
 
 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, Haloalkane dehalogenase 3 | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,3-propandiol
To be Published
|
|
2NU2
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
1SGR
 
 | |
1SGT
 
 | |
