2RQW
 
 | |
2RQV
 
 | |
2RQE
 
 | | Solution structure of the silkworm bGRP/GNBP3 N-terminal domain reveals the mechanism for b-1,3-glucan specific recognition | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Takahasi, K, Ochiai, M, Horiuchi, M, Kumeta, H, Ogura, K, Ashida, M, Inagaki, F. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2RPV
 
 | | Solution Structure of GB1 with LBT probe | | Descriptor: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | Authors: | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
2RMJ
 
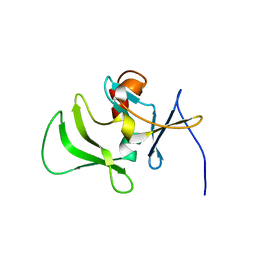 | | Solution structure of RIG-I C-terminal domain | | Descriptor: | Probable ATP-dependent RNA helicase DDX58 | | Authors: | Takahasi, K, Yoneyama, M, Nihishori, T, Hirai, R, Narita, R, Gale Jr, M, Fujita, T, Inagaki, F. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses
Mol.Cell, 29, 2008
|
|
2RSE
 
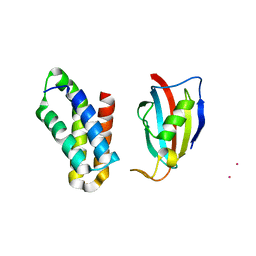 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
1MKC
 
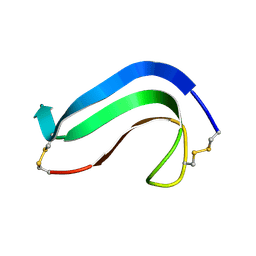 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1Q1O
 
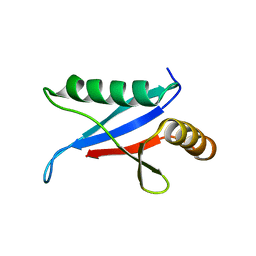 | | Solution Structure of the PB1 Domain of Cdc24p (Long Form) | | Descriptor: | Cell division control protein 24 | | Authors: | Yoshinaga, S, Kohjima, M, Ogura, K, Yokochi, M, Takeya, R, Ito, T, Sumimoto, H, Inagaki, F. | | Deposit date: | 2003-07-22 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PB1 domain and the PC motif-containing region are structurally similar protein binding modules
EMBO J., 22, 2003
|
|
1MKN
 
 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1ERA
 
 | |
1GCQ
 
 | | CRYSTAL STRUCTURE OF VAV AND GRB2 SH3 DOMAINS | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1FRA
 
 | |
1GFD
 
 | |
1GFC
 
 | |
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
1WLP
 
 | | Solution Structure Of The P22Phox-P47Phox Complex | | Descriptor: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | Authors: | Ogura, K, Torikai, S, Saikawa, K, Yuzawa, S, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-06-29 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the tandem Src homology 3 domains of p47phox complexed with a p22phox-derived proline-rich peptide
J.Biol.Chem., 281, 2006
|
|
1WMJ
 
 | |
1WMH
 
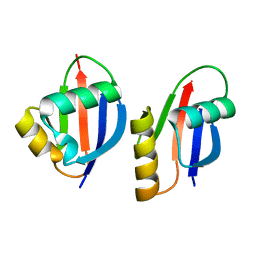 | | Crystal structure of a PB1 domain complex of Protein kinase c iota and Par6 alpha | | Descriptor: | Partitioning defective-6 homolog alpha, Protein kinase C, iota type | | Authors: | Hirano, Y, Yoshinaga, S, Suzuki, N.N, Horiuchi, M, Kohjima, M, Takeya, R, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-07-09 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Cell Polarity Regulator, a Complex between Atypical PKC and Par6 PB1 Domains
J.Biol.Chem., 280, 2005
|
|
1WZ3
 
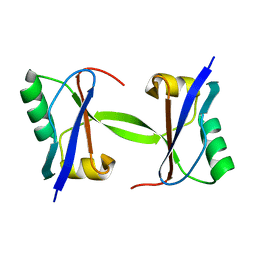 | | The crystal structure of plant ATG12 | | Descriptor: | autophagy 12b | | Authors: | Suzuki, N.N, Yoshimoto, K, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of plant ATG12 and its biological implication in autophagy.
Autophagy, 1, 2005
|
|
1X0N
 
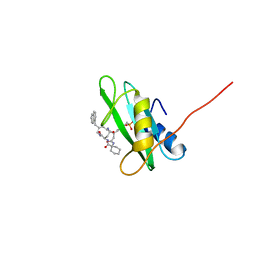 | | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor | | Descriptor: | 4-[(10S,14S,18S)-18-(2-AMINO-2-OXOETHYL)-14-(1-NAPHTHYLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL]BENZYLPHOSPHONIC ACID, Growth factor receptor-bound protein 2 | | Authors: | Ogura, K, Shiga, T, Yuzawa, S, Yokochi, M, Burke, T.R, Inagaki, F. | | Deposit date: | 2005-03-24 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of growth factor receptor binding protein SH2 domain complexed with the inhibitor
To be Published
|
|
1WLZ
 
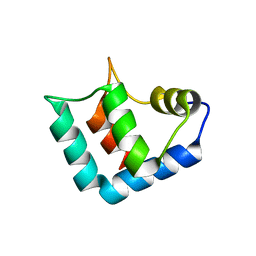 | | Crystal structure of DJBP fragment which was obtained by limited proteolysis | | Descriptor: | CAP-binding protein complex interacting protein 1 isoform a | | Authors: | Honbou, K, Suzuki, N, Horiuchi, M, Taira, T, Niki, T, Ariga, H, Inagaki, F. | | Deposit date: | 2004-07-01 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DJBP Fragment which was obtained by Limited Proteolysis
To be Published
|
|
1VD2
 
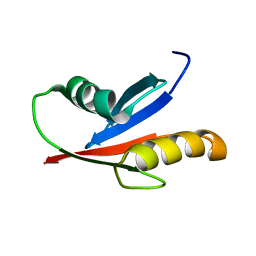 | | Solution Structure of the PB1 domain of PKCiota | | Descriptor: | Protein kinase C, iota type | | Authors: | Hirano, Y, Yoshinaga, S, Yokochi, M, Ogura, K, Noda, Y, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-03-18 | | Release date: | 2004-09-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5
J.Biol.Chem., 279, 2004
|
|
1Z9Q
 
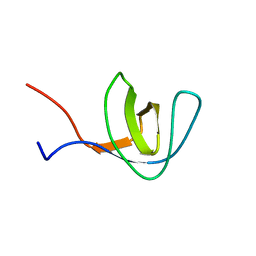 | |
2DYM
 
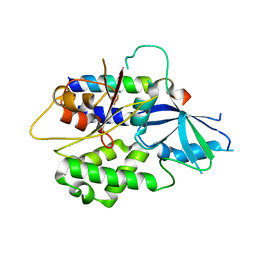 | |
2DYT
 
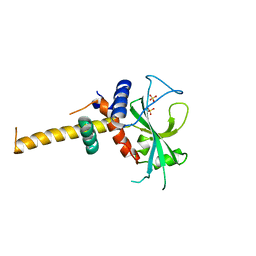 | |
