5CWW
 
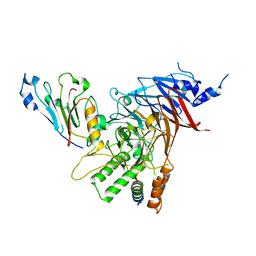 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
7AJU
 
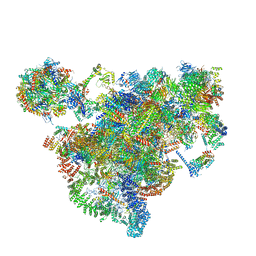 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7AJT
 
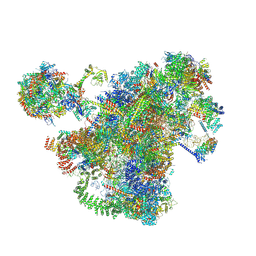 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
6ZQF
 
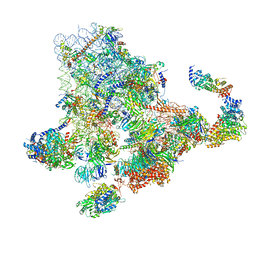 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-B (Poly-Ala) | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQB
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state B2 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQD
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
4GYG
 
 | | Crystal structure of the Rio2 kinase from Chaetomium thermophilum | | Descriptor: | Rio2 kinase | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GYI
 
 | | Crystal structure of the Rio2 kinase-ADP/Mg2+-phosphoaspartate complex from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6EM5
 
 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6QTB
 
 | | Crystal structure of Rea1-MIDAS/Ytm1-UBL complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, MAGNESIUM ION, Midasin,Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
6RXU
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1 | | Descriptor: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXY
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state a | | Descriptor: | 35S rRNA, 40S ribosomal protein S13-like protein, 40S ribosomal protein S14-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXX
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state C, Poly-Ala | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXV
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6QT8
 
 | | Crystal structure of Rea1-MIDAS domain from Chaetomium thermophilum | | Descriptor: | GLYCEROL, IODIDE ION, Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
6RXZ
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S11-like protein, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXT
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
1AC9
 
 | | SOLUTION STRUCTURE OF A DNA DECAMER CONTAINING THE ANTIVIRAL DRUG GANCICLOVIR: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS, AND FULL RELAXATION REFINEMENT, 6 STRUCTURES | | Descriptor: | DNA | | Authors: | Foti, M, Marshalko, S, Schurter, E, Kumar, S, Beardsley, G.P, Schweitzer, B.I. | | Deposit date: | 1997-02-17 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer containing the antiviral drug ganciclovir: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 36, 1997
|
|
3KJL
 
 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1.
J.Biol.Chem., 285, 2010
|
|
2Q8K
 
 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
5OOQ
 
 | | Structure of the Mtr4 Nop53 Complex | | Descriptor: | ATP-dependent RNA helicase DOB1, Ribosome biogenesis protein NOP53, SULFATE ION | | Authors: | Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the preribosomal protein Nop53.
RNA, 23, 2017
|
|
3KIK
 
 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1
J.Biol.Chem., 285, 2010
|
|
4OTP
 
 | |
4NWB
 
 | | Crystal structure of Mrt4 | | Descriptor: | SULFATE ION, mRNA turnover protein 4 | | Authors: | Holdermann, I, Sinning, I. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
3FWB
 
 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1 | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
