4E7K
 
 | | PFV integrase Target Capture Complex (TCC-Mn), freeze-trapped prior to strand transfer, at 3.0 A resolution | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*GP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*CP*TP*CP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maertens, G.N, Cherepanov, P. | | Deposit date: | 2012-03-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | 3'-Processing and strand transfer catalysed by retroviral integrase in crystallo.
Embo J., 31, 2012
|
|
8OUD
 
 | |
8AXP
 
 | |
5COF
 
 | | Crystal structure of Uncharacterised protein Q1R1X2 from Escherichia coli UTI89 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Taylor, J.D, Hare, S, Matthews, S.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5CQV
 
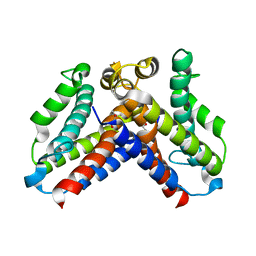 | | Crystal structure of uncharacterized protein Q8DWV2 from Streptococcus agalactiae | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Uncharacterized protein | | Authors: | Taylor, J.D, Hare, S, Matthews, S.J. | | Deposit date: | 2015-07-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
3DLR
 
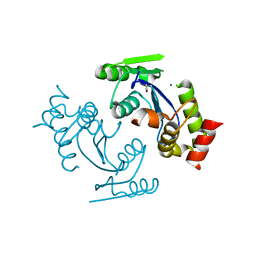 | |
4C0O
 
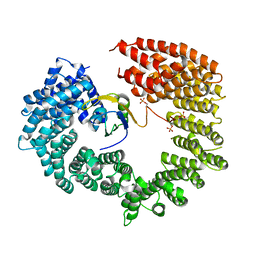 | |
4IWW
 
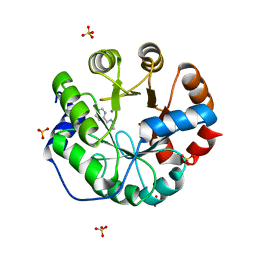 | | Computational Design of an Unnatural Amino Acid Metalloprotein with Atomic Level Accuracy | | Descriptor: | COBALT (II) ION, SULFATE ION, Unnatural Amino Acid Mediated Metalloprotein | | Authors: | Mills, J, Bolduc, J, Khare, S, Stoddard, B, Baker, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
4IX0
 
 | | Computational Design of an Unnatural Amino Acid Metalloprotein with Atomic Level Accuracy | | Descriptor: | NICKEL (II) ION, SULFATE ION, Unnatural Amino Acid Mediated Metalloprotein | | Authors: | Mills, J, Bolduc, J, Khare, S, Stoddard, B, Baker, D. | | Deposit date: | 2013-01-24 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
8D8Z
 
 | | Crystal structure of ChoE N147A mutant in complex with thiocholine and chloride | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, CHLORIDE ION, ChoE, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D91
 
 | | Crystal structure of ChoE in complex with acetate and tetraethylammonium (TEA) | | Descriptor: | ACETATE ION, ChoE, TETRAETHYLAMMONIUM ION | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8W
 
 | | Crystal structure of ChoE with Ser38 adopting alternative conformations | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ChoE, IODIDE ION | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8Y
 
 | | Crystal structure of ChoE N147A mutant in complex with acetylthiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ACETYLTHIOCHOLINE, CHLORIDE ION, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D90
 
 | | Crystal structure of ChoE N147A mutant in complex with bromide ions | | Descriptor: | BROMIDE ION, ChoE, GLYCEROL | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8X
 
 | | Crystal structure of ChoE in complex with acetate and thiocholine (crystal form 2) | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
5M4V
 
 | | X-ray structure of the mambaquaretin-1, a selective antagonist of the vasopressin type 2 receptor | | Descriptor: | CHLORIDE ION, Mambaquaretin-1, S-1,2-PROPANEDIOL | | Authors: | Stura, E.A, Vera, L, Ciolek, J, Mourier, G, Gilles, N. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8GLT
 
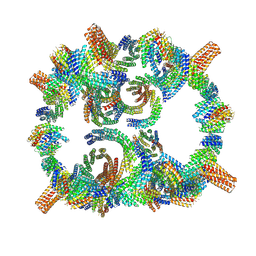 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | Descriptor: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | Authors: | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 20, 2024
|
|
8EVM
 
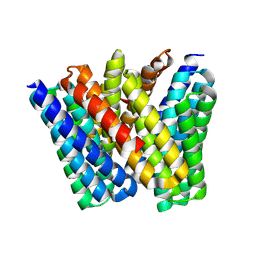 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
8SXP
 
 | |
7UNJ
 
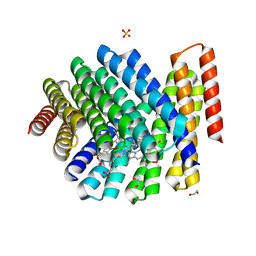 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UKR
 
 | | Crystal Structure of SOS1 with MRTX0902, a Potent and Selective Inhibitor of the SOS1:KRAS Protein-Protein Interaction | | Descriptor: | 2-methyl-3-[(1R)-1-{[4-methyl-7-(morpholin-4-yl)pyrido[3,4-d]pyridazin-1-yl]amino}ethyl]benzonitrile, Son of sevenless homolog 1 | | Authors: | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7UKS
 
 | | Crystal structure of SOS1 with phthalazine inhibitor bound (compound 15) | | Descriptor: | 4-methyl-N-{(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}-7-(piperazin-1-yl)phthalazin-1-amine, Son of sevenless homolog 1 | | Authors: | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
4QTF
 
 | |
