2N64
 
 | |
2M97
 
 | |
2KDU
 
 | | Structural basis of the Munc13-1/Ca2+-Calmodulin interaction: A novel 1-26 calmodulin binding motif with a bipartite binding mode | | Descriptor: | CALCIUM ION, Calmodulin, Protein unc-13 homolog A | | Authors: | Rodriguez-Castaneda, F.A, Maestre-Martinez, M, Coudevylle, N, Dimova, K, Jahn, O, Junge, H, Becker, S, Brose, N, Carlomagno, T, Griesinger, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity.
Embo J., 29, 2010
|
|
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2KOX
 
 | | NMR residual dipolar couplings identify long range correlated motions in the backbone of the protein ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Fenwick, R.B, Richter, B, Lee, D, Walter, K.F.A, Milovanovic, D, Becker, S, Lakomek, N.A, Griesinger, C, Salvatella, X. | | Deposit date: | 2009-10-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Weak Long-Range Correlated Motions in a Surface Patch of Ubiquitin Involved in Molecular Recognition
J.Am.Chem.Soc., 2011
|
|
1HXV
 
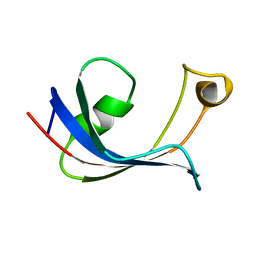 | | PPIASE DOMAIN OF THE MYCOPLASMA GENITALIUM TRIGGER FACTOR | | Descriptor: | TRIGGER FACTOR | | Authors: | Vogtherr, M, Parac, T.N, Maurer, M, Pahl, A, Fiebig, K. | | Deposit date: | 2001-01-17 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and dynamics of the peptidyl-prolyl cis-trans isomerase domain of the trigger factor from Mycoplasma genitalium compared to FK506-binding protein.
J.Mol.Biol., 318, 2002
|
|
1X6M
 
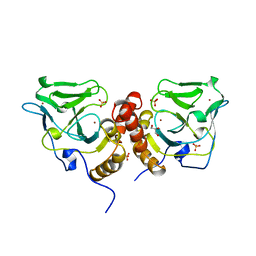 | | Crystal structure of the glutathione-dependent formaldehyde-activating enzyme (Gfa) | | Descriptor: | GLYCEROL, Glutathione-dependent formaldehyde-activating enzyme, SULFATE ION, ... | | Authors: | Neculai, A.M, Neculai, D, Vorholt, J.A, Becker, S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A dynamic zinc redox switch
J.Biol.Chem., 280, 2005
|
|
7QI2
 
 | |
7OOJ
 
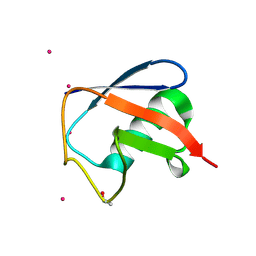 | | Structure of D-Thr53 Ubiquitin | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Becker, S. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A litmus test for classifying recognition mechanisms of transiently binding proteins.
Nat Commun, 13, 2022
|
|
2KUZ
 
 | | 2-Aminopurine incorporation perturbs the dynamics and structure of DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*TP*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*AP*AP*CP*GP*TP*CP*G)-3') | | Authors: | Dallmann, A, Dehmel, L, Peters, T, Muegge, C, Griesinger, C.P, Tuma, J, Ernsting, N.P. | | Deposit date: | 2010-03-03 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 2-aminopurine incorporation perturbs the dynamics and structure of DNA.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2KV0
 
 | | 2-Aminopurine incorporation perturbs the dynamics and structure of DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*TP*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*(2PR)P*AP*CP*GP*TP*CP*G)-3') | | Authors: | Dallmann, A, Dehmel, L, Peters, T, Muegge, C, Griesinger, C.P, Tuma, J, Ernsting, N.P. | | Deposit date: | 2010-03-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 2-Aminopurine incorporation perturbs the dynamics and structure of DNA.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3DNE
 
 | | cAMP-dependent protein kinase PKA catalytic subunit with PKI-5-24 | | Descriptor: | 3-pyridin-4-yl-1H-indazole, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2008-07-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallography-independent determination of ligand binding modes
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
5FQ1
 
 | |
3DND
 
 | | cAMP-dependent protein kinase PKA catalytic subunit with PKI-5-24 | | Descriptor: | 5-benzyl-1,3-thiazol-2-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2008-07-02 | | Release date: | 2009-06-23 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallography-independent determination of ligand binding modes
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
4YXS
 
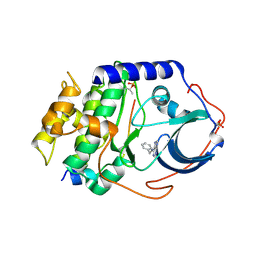 | | CAMP-DEPENDENT PROTEIN KINASE PKA CATALYTIC SUBUNIT WITH PKI-5-24 | | Descriptor: | N-BENZYL-9H-PURIN-6-AMINE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-20 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4YXR
 
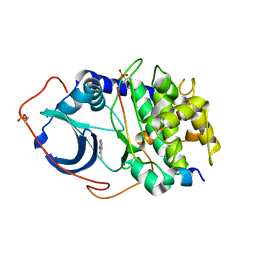 | | CRYSTAL STRUCTURE OF PKA IN COMPLEX WITH inhibitor. | | Descriptor: | 3-methyl-2H-indazole, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2LEV
 
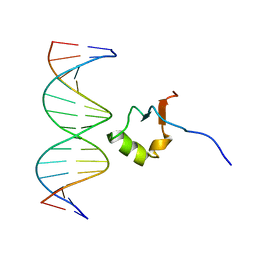 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
1YGW
 
 | |
2LGC
 
 | |
2KN5
 
 | |
