4RQZ
 
 | |
4RR0
 
 | |
4RQY
 
 | |
4RR1
 
 | |
1G2D
 
 | | STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (CLONE #2) | | Descriptor: | 5'-D(*GP*AP*CP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*AP*G)-3', 5'-D(*TP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*GP*TP*CP*C)-3', TATA BOX ZINC FINGER PROTEIN, ... | | Authors: | Wolfe, S.A, Grant, R.A, Elrod-Erickson, M, Pabo, C.O. | | Deposit date: | 2000-10-18 | | Release date: | 2001-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Beyond the "recognition code": structures of two Cys2His2 zinc finger/TATA box complexes.
Structure, 9, 2001
|
|
1YFN
 
 | | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB- the crystal structure of a SspB-RseA complex | | Descriptor: | Sigma-E factor negative regulatory protein, Stringent starvation protein B | | Authors: | Levchenko, I, Grant, R.A, Flynn, J.M, Sauer, R.T, Baker, T.A. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YWT
 
 | | Crystal structure of the human sigma isoform of 14-3-3 in complex with a mode-1 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, synthetic optimal phosphopeptide (mode-1) | | Authors: | Wilker, E.W, Grant, R.A, Artim, S.C, Yaffe, M.B. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for 14-3-3sigma functional specificity.
J.Biol.Chem., 280, 2005
|
|
1ZSZ
 
 | | Crystal structure of a computationally designed SspB heterodimer | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2005-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity versus stability in computational protein design.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TWB
 
 | | SspB disulfide crosslinked to an ssrA degradation tag | | Descriptor: | Stringent starvation protein B homolog, ssrA peptide | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide-Dependent Substrate Handoff from the SspB Adaptor to the AAA+ ClpXP Protease.
Mol.Cell, 16, 2004
|
|
1U9P
 
 | | Permuted single-chain Arc | | Descriptor: | pArc | | Authors: | Tabtiang, R.K, Cezairliyan, B.O, Grant, R.A, Cochrane, J.C, Sauer, R.T. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Consolidating critical binding determinants by noncyclic rearrangement of protein secondary structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1LLM
 
 | |
1OU8
 
 | | structure of an AAA+ protease delivery protein in complex with a peptide degradation tag | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog, synthetic ssrA peptide | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1OU9
 
 | | Structure of SspB, a AAA+ protease delivery protein | | Descriptor: | CALCIUM ION, Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
1OUL
 
 | | Structure of the AAA+ protease delivery protein SspB | | Descriptor: | Stringent starvation protein B homolog | | Authors: | Levchenko, I, Grant, R.A, Wah, D.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a delivery protein for an AAA+ protease in complex with a peptide degradation tag
Mol.Cell, 12, 2003
|
|
6VWO
 
 | | Crystal structure of E. coli guanosine kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE, Inosine-guanosine kinase, ... | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation.
Mol.Cell, 80, 2020
|
|
6VWP
 
 | | Crystal structure of E. coli guanosine kinase in complex with ppGpp | | Descriptor: | GUANOSINE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-guanosine kinase, ... | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | ppGpp Coordinates Nucleotide and Amino-Acid Synthesis in E. coli During Starvation.
Mol.Cell, 80, 2020
|
|
6X0A
 
 | | X-ray structure of a chimeric ParDE toxin-antitoxin complex from Mesorhizobium opportunistum | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Plasmid stabilization system, Putative addiction module antidote protein, ... | | Authors: | Lite, T.L, Grant, R.A, Laub, M.T. | | Deposit date: | 2020-05-15 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library.
Elife, 9, 2020
|
|
3O2H
 
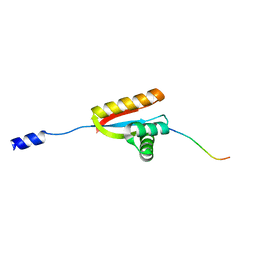 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
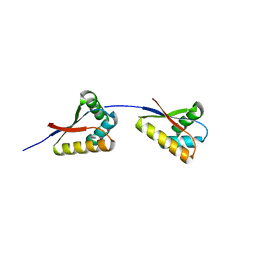 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
1F2I
 
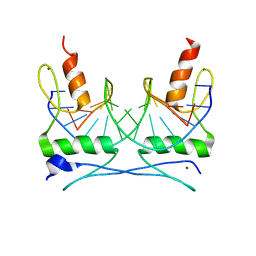 | | COCRYSTAL STRUCTURE OF SELECTED ZINC FINGER DIMER BOUND TO DNA | | Descriptor: | 5'-D(*AP*TP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*AP*T)-3', FUSION OF N-TERMINAL 17-MER PEPTIDE EXTENSION TO ZIF12, ZINC ION | | Authors: | Wang, B.S, Grant, R.A, Pabo, C.O. | | Deposit date: | 2000-05-25 | | Release date: | 2001-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Selected peptide extension contacts hydrophobic patch on neighboring zinc finger and mediates dimerization on DNA.
Nat.Struct.Biol., 8, 2001
|
|
3ES2
 
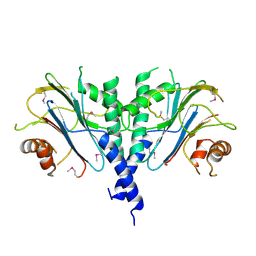 | |
3EOD
 
 | |
3OTP
 
 | |
3DNJ
 
 | | The structure of the Caulobacter crescentus ClpS protease adaptor protein in complex with a N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, synthetic N-end rule peptide | | Authors: | Wang, K, Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2008-07-02 | | Release date: | 2008-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The molecular basis of N-end rule recognition.
Mol.Cell, 32, 2008
|
|
3F7A
 
 | |
